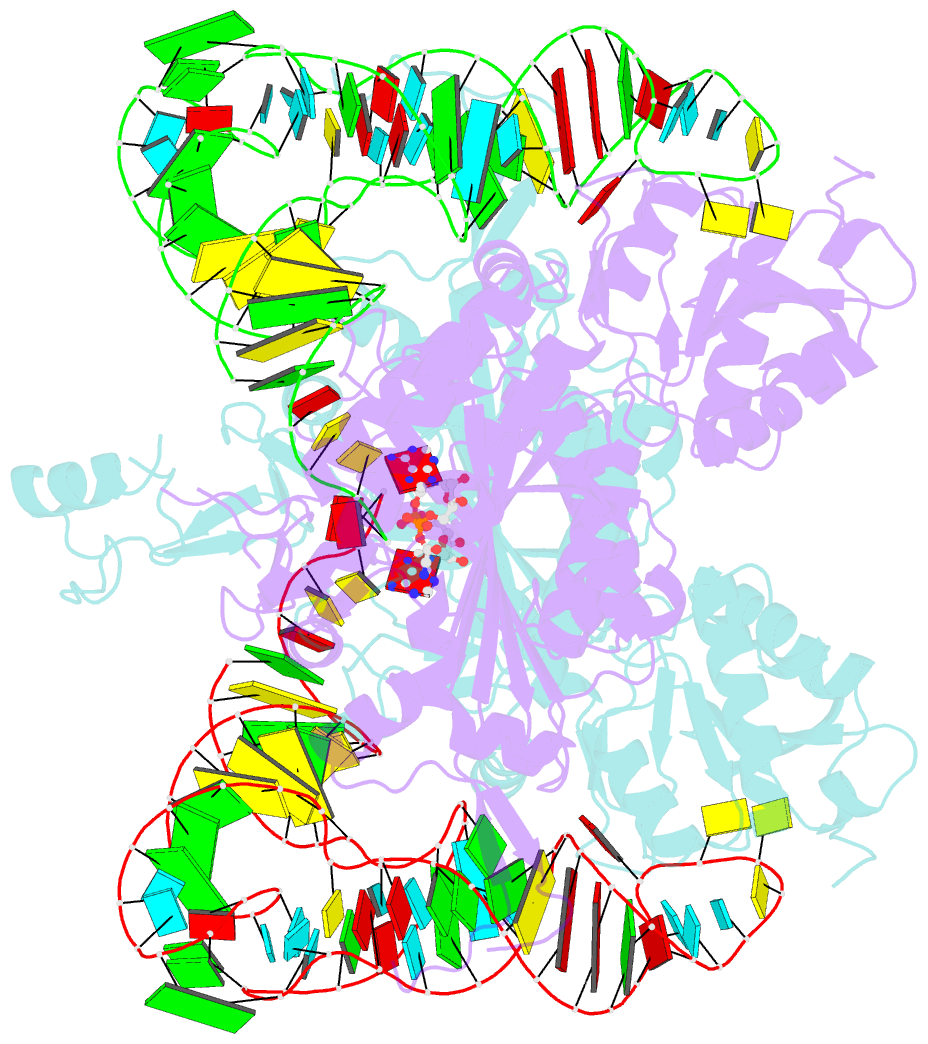
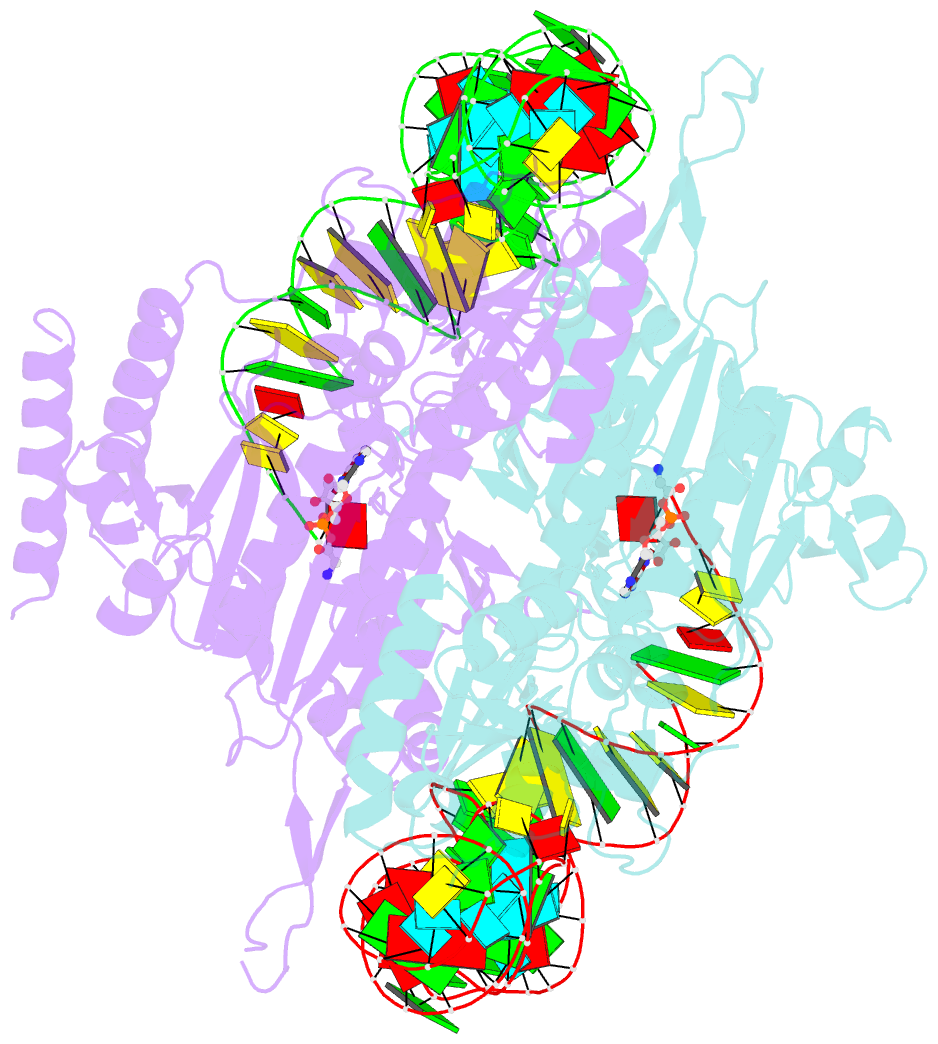
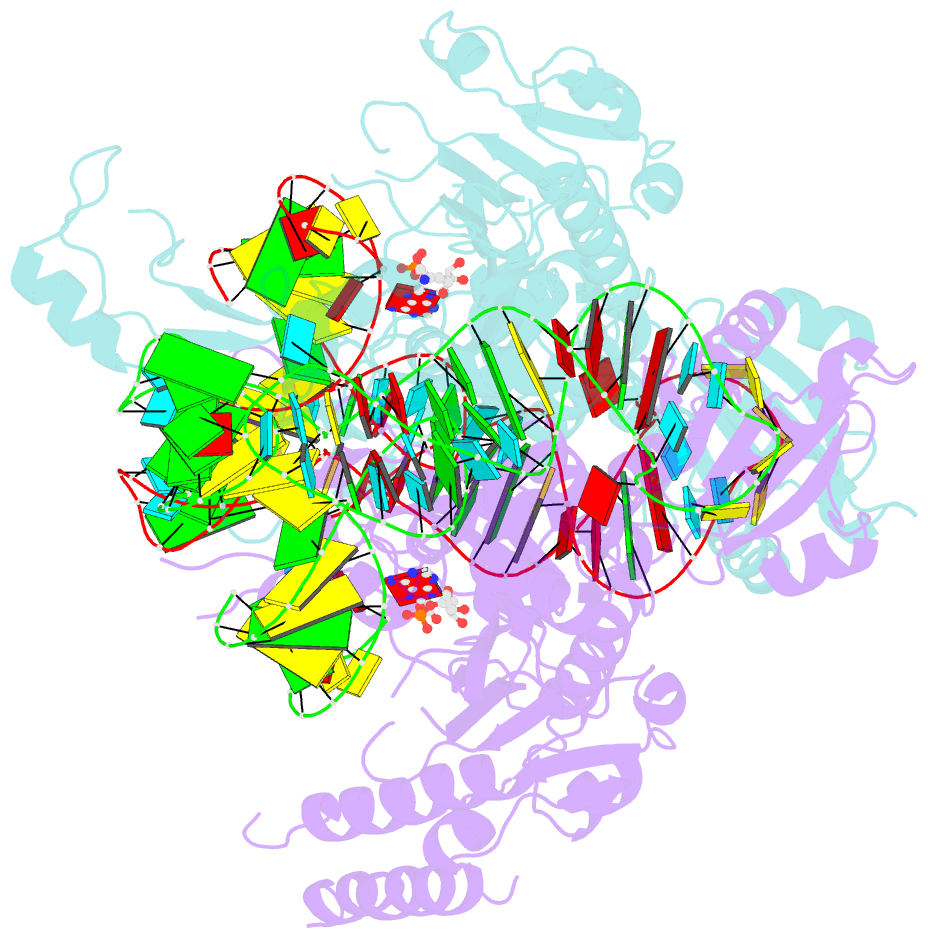
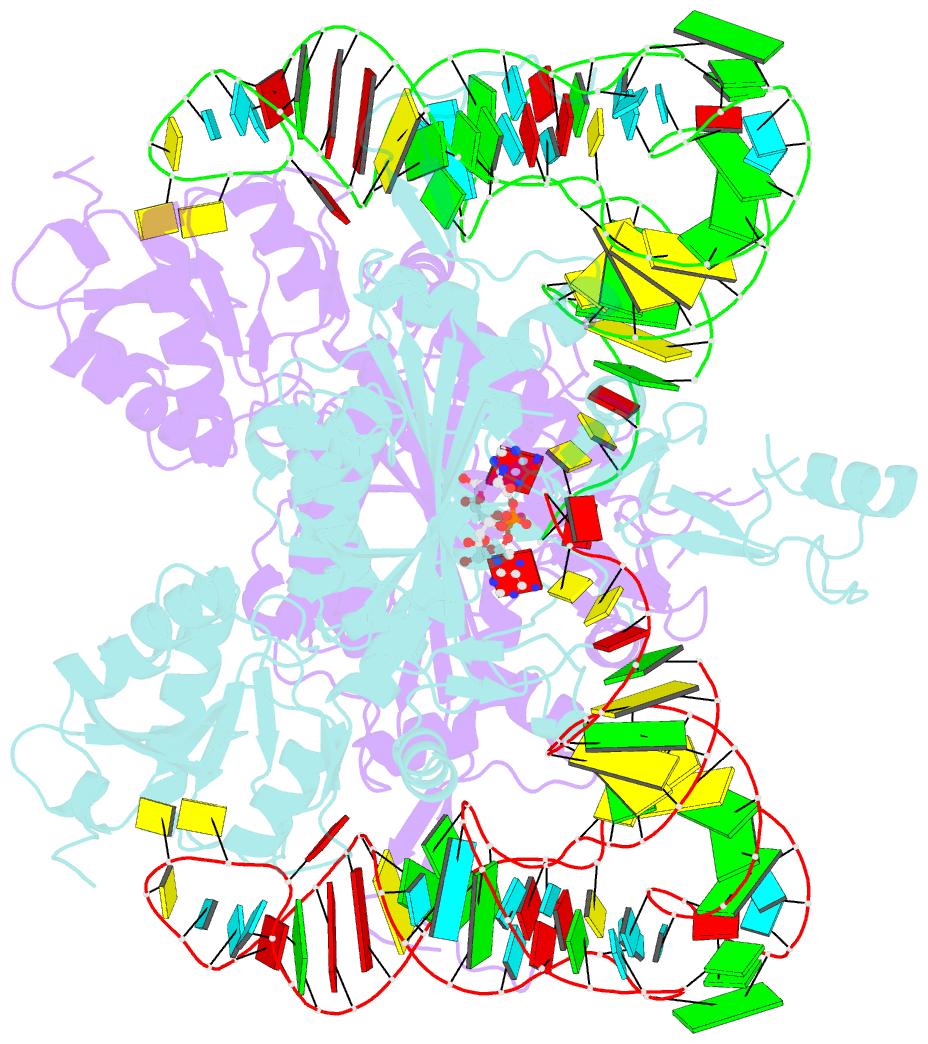
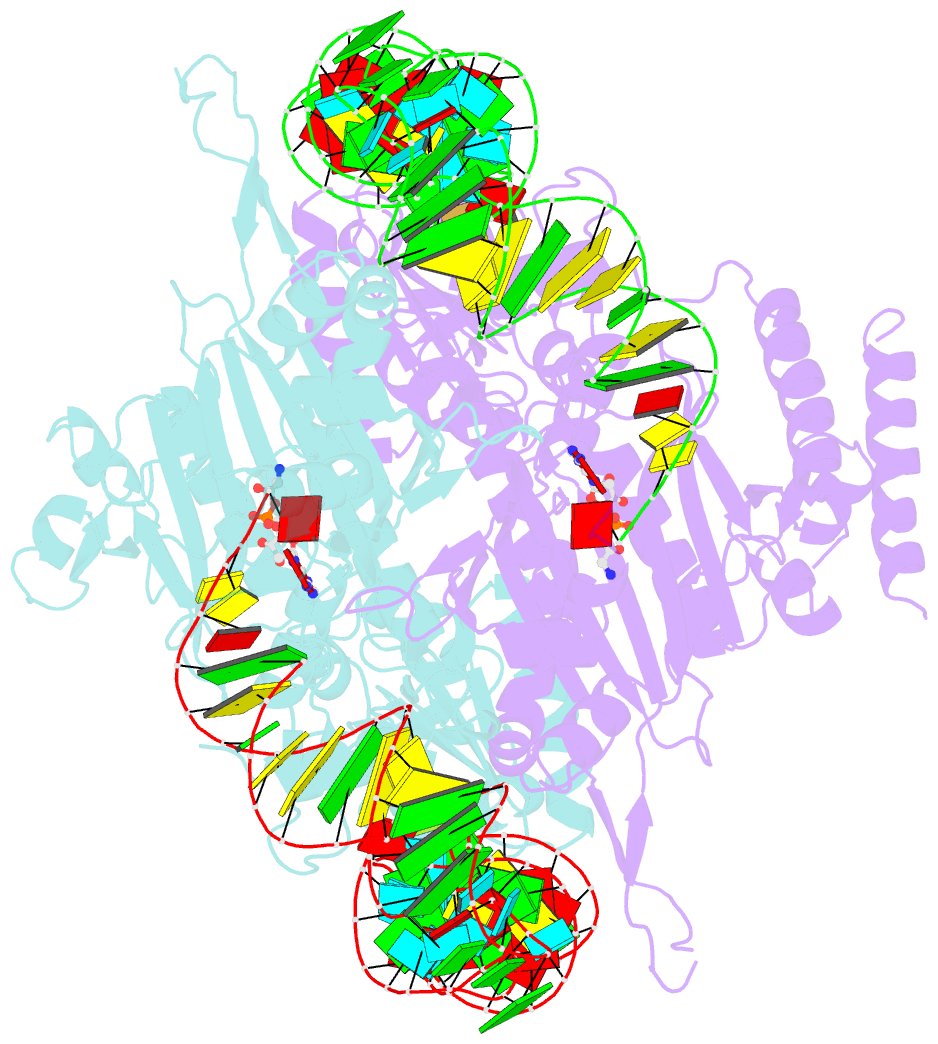
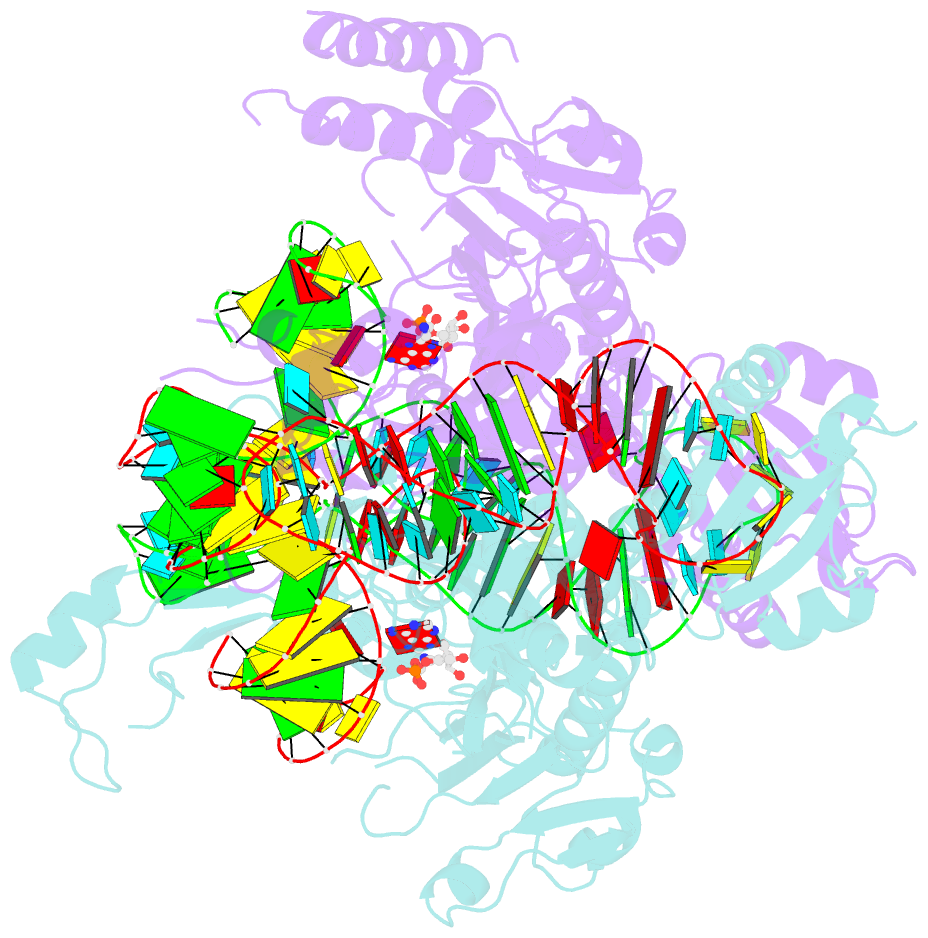
Summary information and primary citation
- PDB-id
- 5e6m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (2.927 Å)
- Summary
- Crystal structure of human wild type glyrs bound with trnagly
- Reference
- Qin X, Deng X, Chen L, Xie W (2016): "Crystal Structure of the Wild-Type Human GlyRS Bound with tRNA(Gly) in a Productive Conformation." J.Mol.Biol., 428, 3603-3614. doi: 10.1016/j.jmb.2016.05.018.
- Abstract
- Aminoacyl-tRNA synthetases are essential components of the protein translational machinery in all living species, among which the human glycyl-tRNA synthetase (hGlyRS) is of great research interest because of its unique species-specific aminoacylation properties and noncanonical roles in the Charcot-Marie-Tooth neurological disease. However, the molecular mechanisms of how the enzyme carries out its classical and alternative functions are not well understood. Here, we report a complex structure of the wild-type hGlyRS bound with tRNA(Gly) at 2.95Å. In the complex, the flexible Whep-TRS domain is visible in one of the subunits of the enzyme dimer, and the tRNA molecule is also completely resolved. At the active site, a glycyl-AMP molecule is synthesized and is waiting for the transfer of the glycyl moiety to occur. This cocrystal structure provides us with new details about the recognition mechanism in the intermediate stage during glycylation, which was not well elucidated in the previous crystal structures where the inhibitor AMPPNP was used for crystallization. More importantly, the structural and biochemical work conducted in the current and previous studies allows us to build a model of the full-length hGlyRS in complex with tRNA(Gly), which greatly helps us to understand the roles that insertions and the Whep-TRS domain play in the tRNA-binding process. Finally, through structure comparison with other class II aminoacyl-tRNA synthetases bound with their tRNA substrates, we found some commonalities of the aminoacylation mechanism between these enzymes.