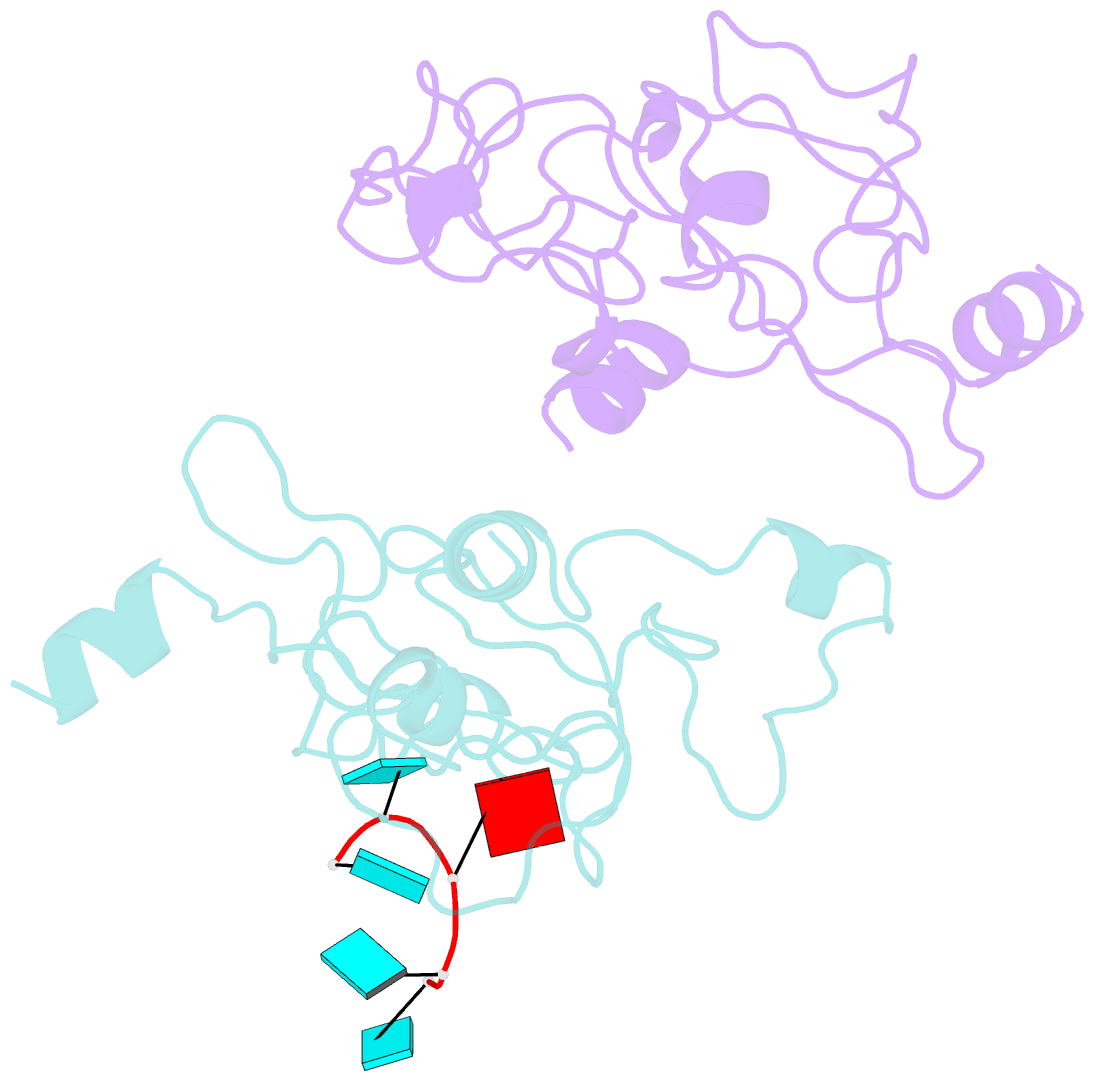
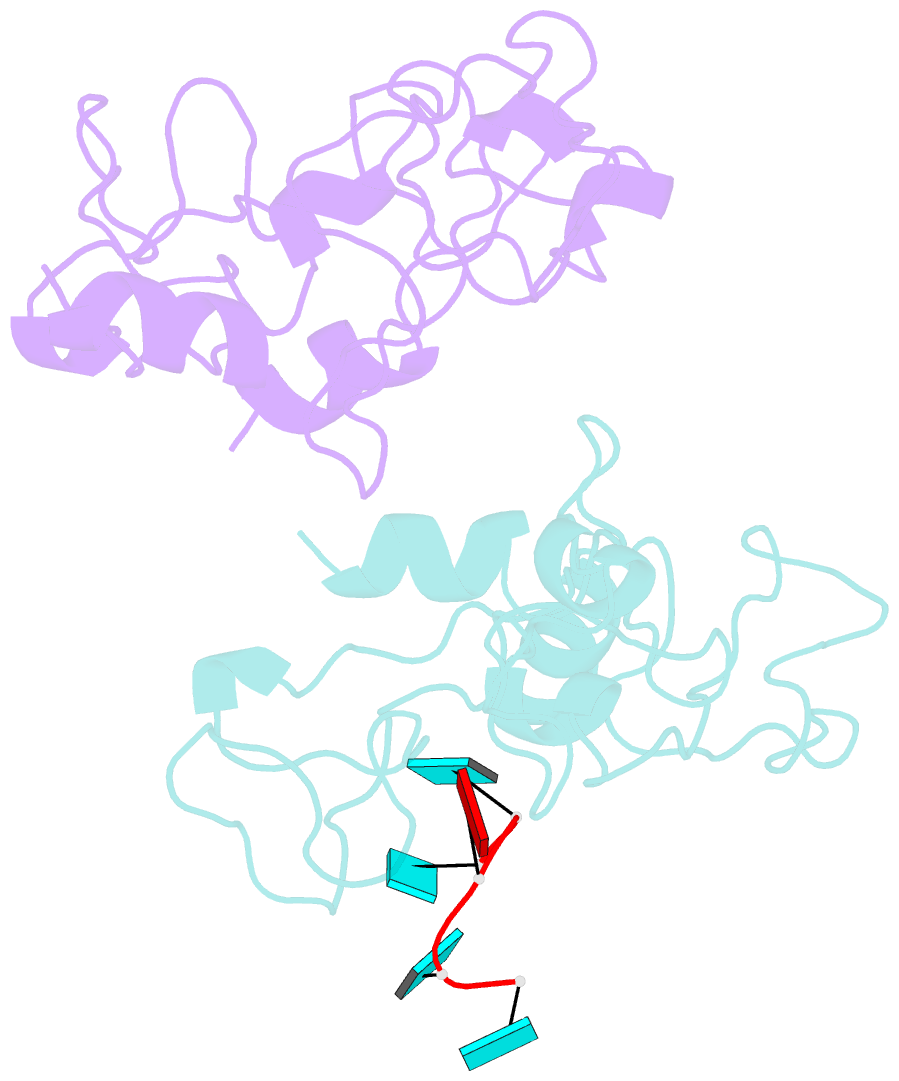
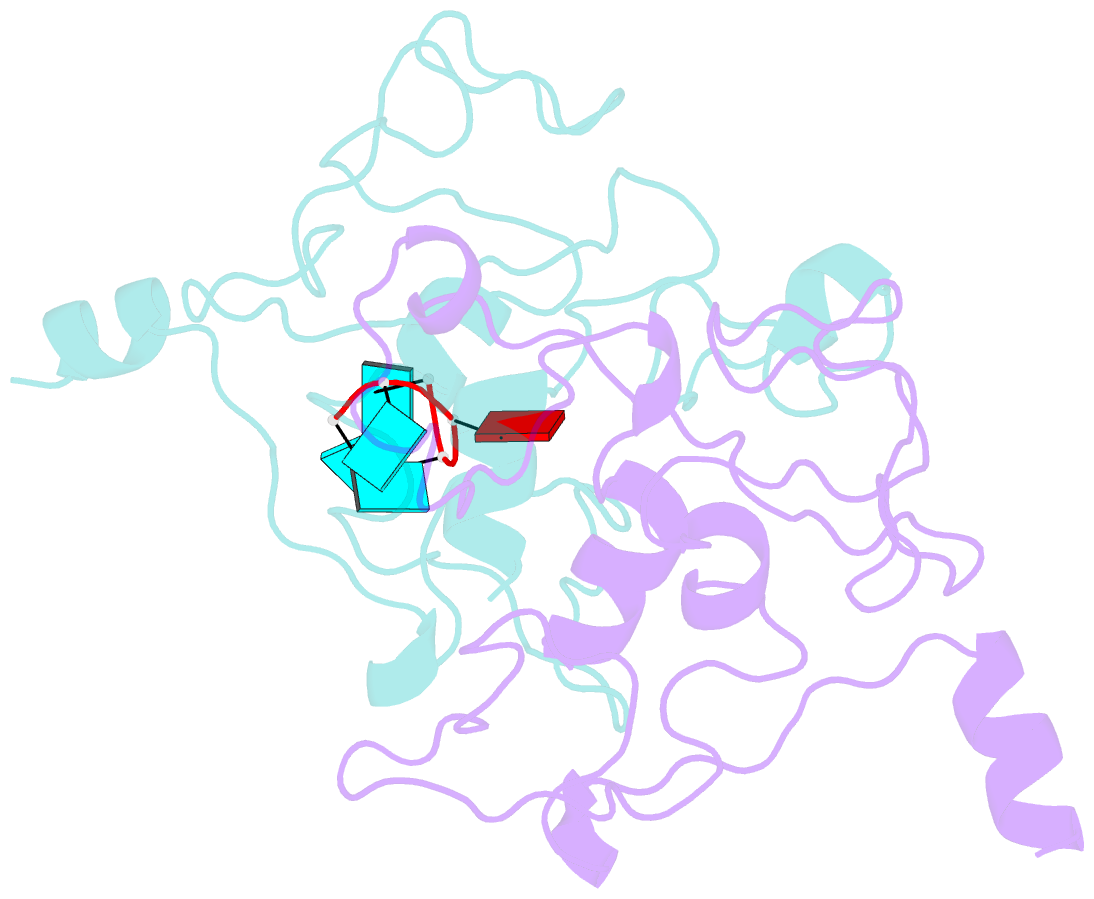
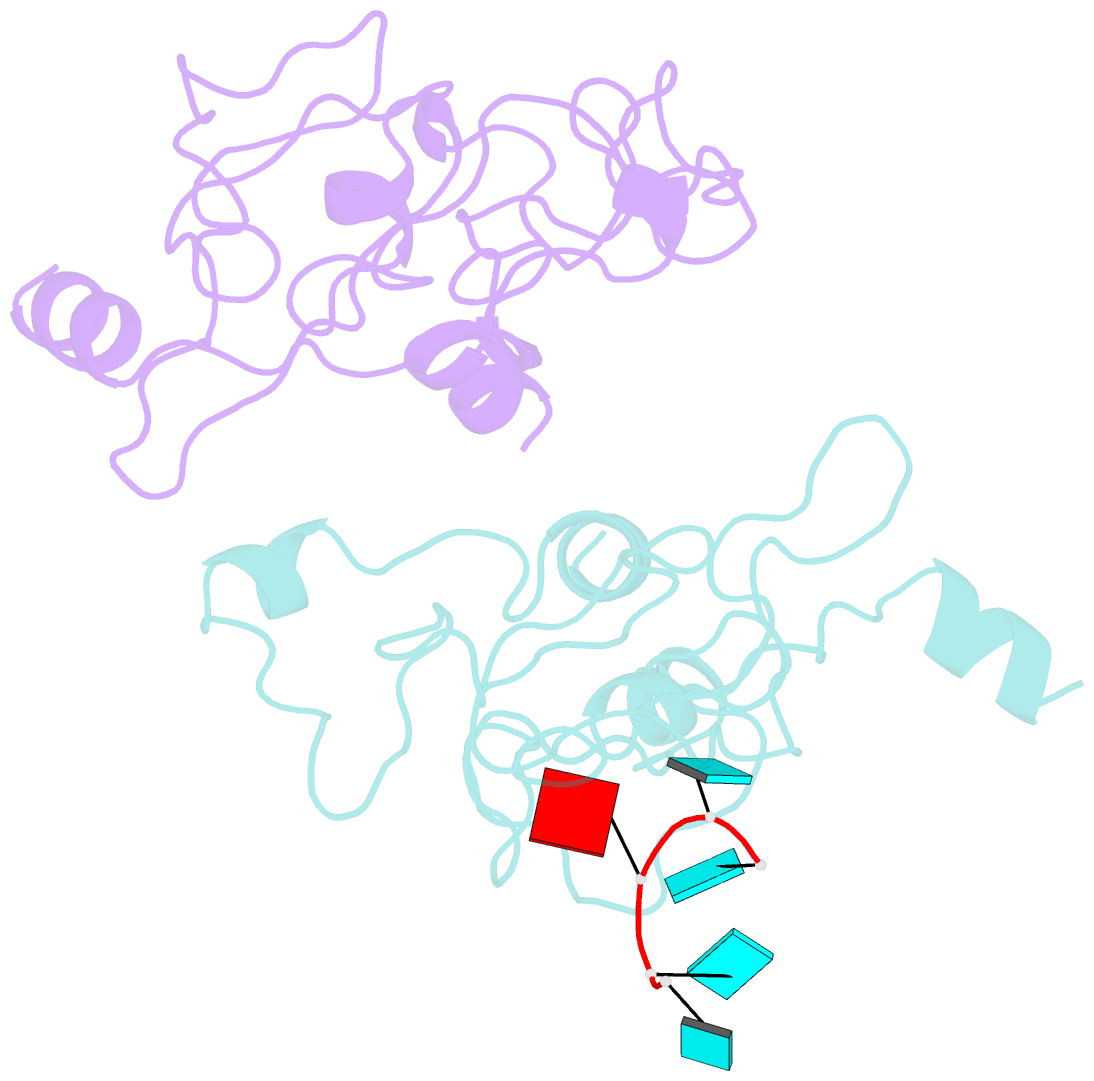
Summary information and primary citation
- PDB-id
- 5elh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (1.8 Å)
- Summary
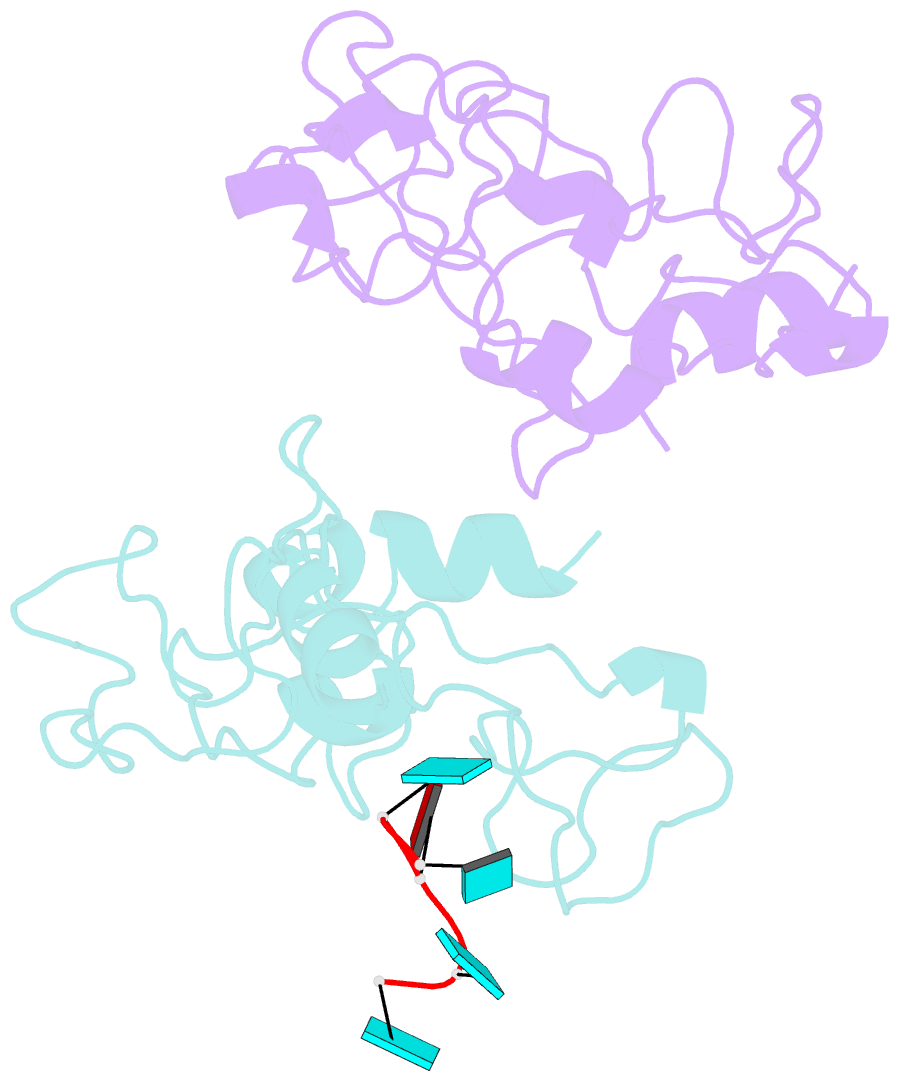
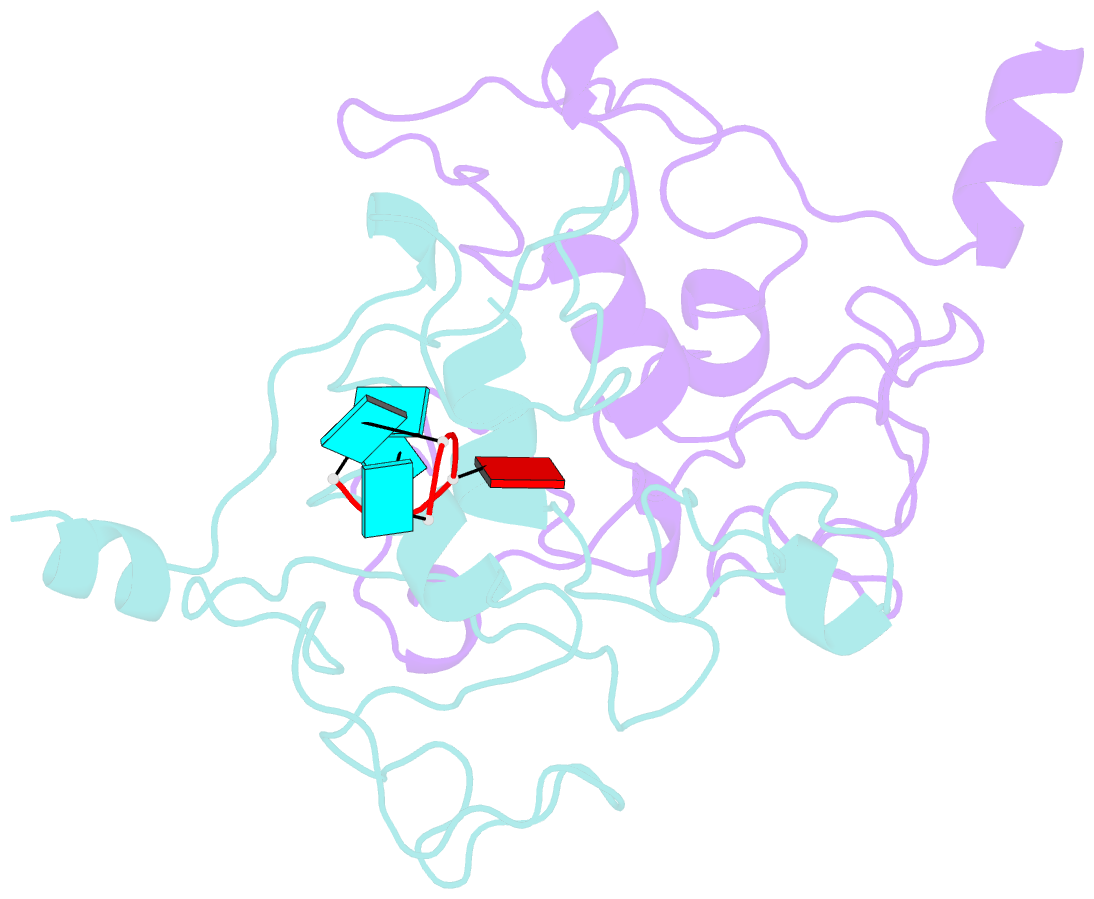
- Crystal structure of mouse unkempt zinc fingers 1-3 (znf1-3), bound to RNA
- Reference
- Murn J, Teplova M, Zarnack K, Shi Y, Patel DJ (2016): "Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt." Nat.Struct.Mol.Biol., 23, 16-23. doi: 10.1038/nsmb.3140.
- Abstract
- Unkempt is an evolutionarily conserved RNA-binding protein that regulates translation of its target genes and is required for the establishment of the early bipolar neuronal morphology. Here we determined the X-ray crystal structure of mouse Unkempt and show that its six CCCH zinc fingers (ZnFs) form two compact clusters, ZnF1-3 and ZnF4-6, that recognize distinct trinucleotide RNA substrates. Both ZnF clusters adopt a similar overall topology and use distinct recognition principles to target specific RNA sequences. Structure-guided point mutations reduce the RNA binding affinity of Unkempt both in vitro and in vivo, ablate Unkempt's translational control and impair the ability of Unkempt to induce a bipolar cellular morphology. Our study unravels a new mode of RNA sequence recognition by clusters of CCCH ZnFs that is critical for post-transcriptional control of neuronal morphology.