Summary information and primary citation
- PDB-id
- 5els; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- X-ray (2.873 Å)
- Summary
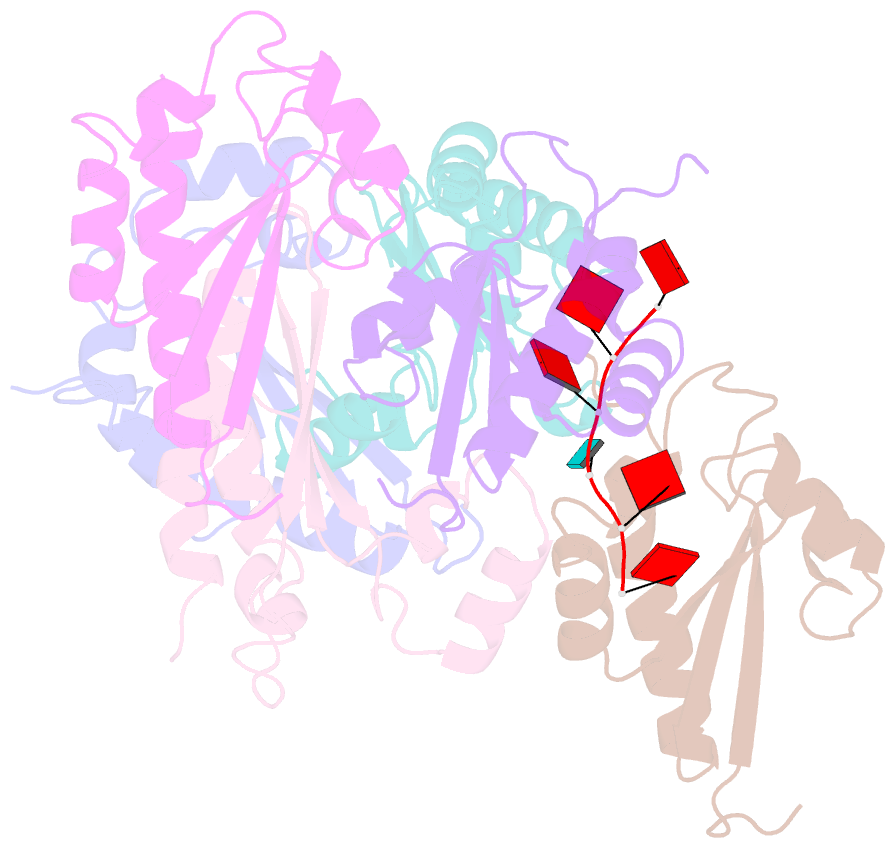
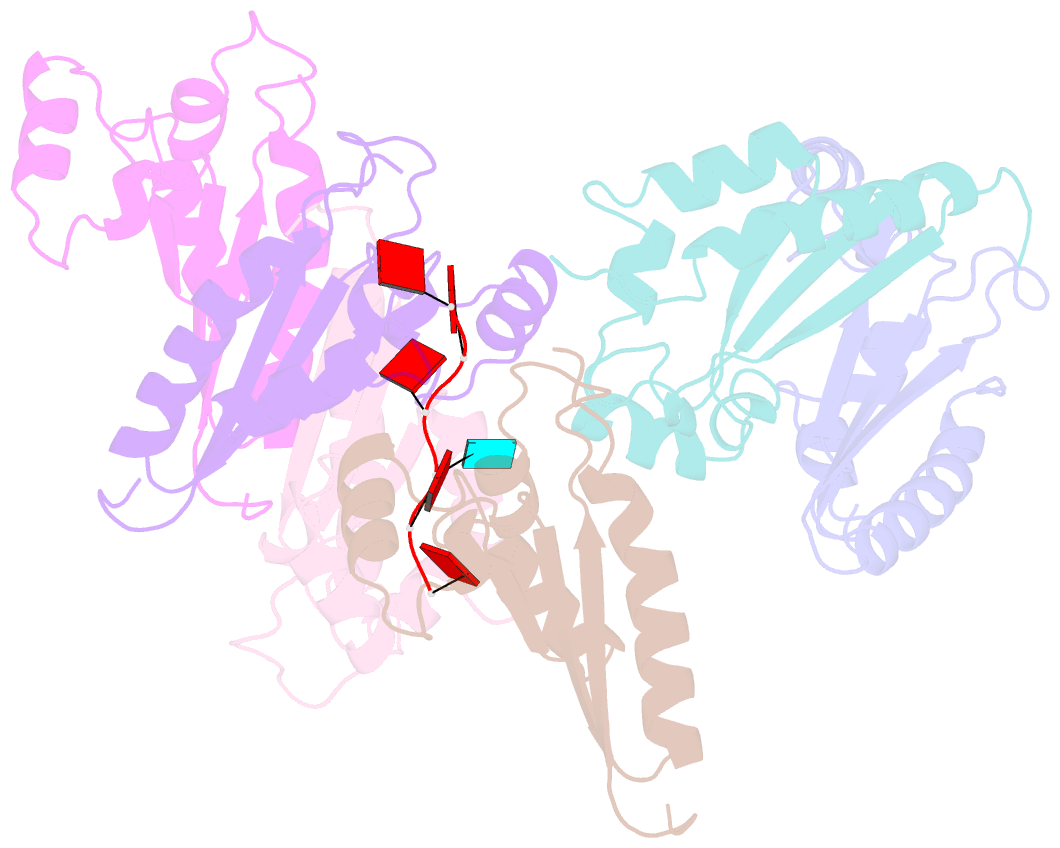
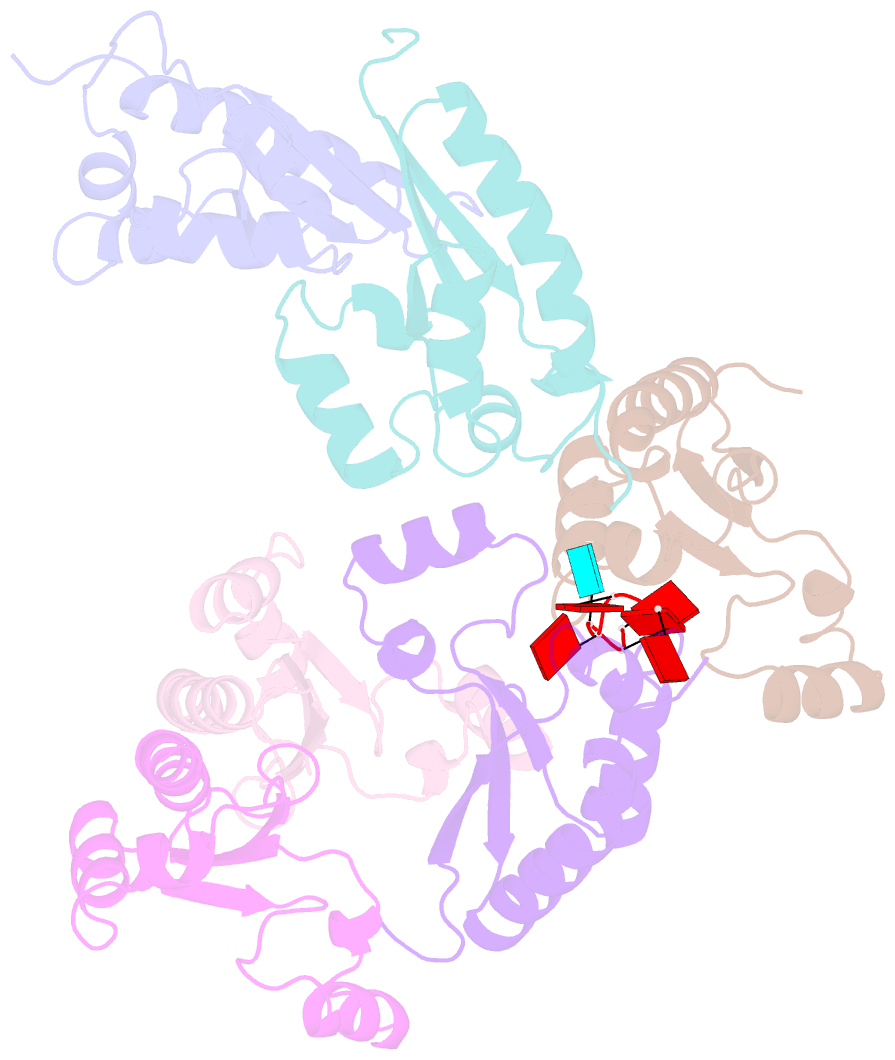
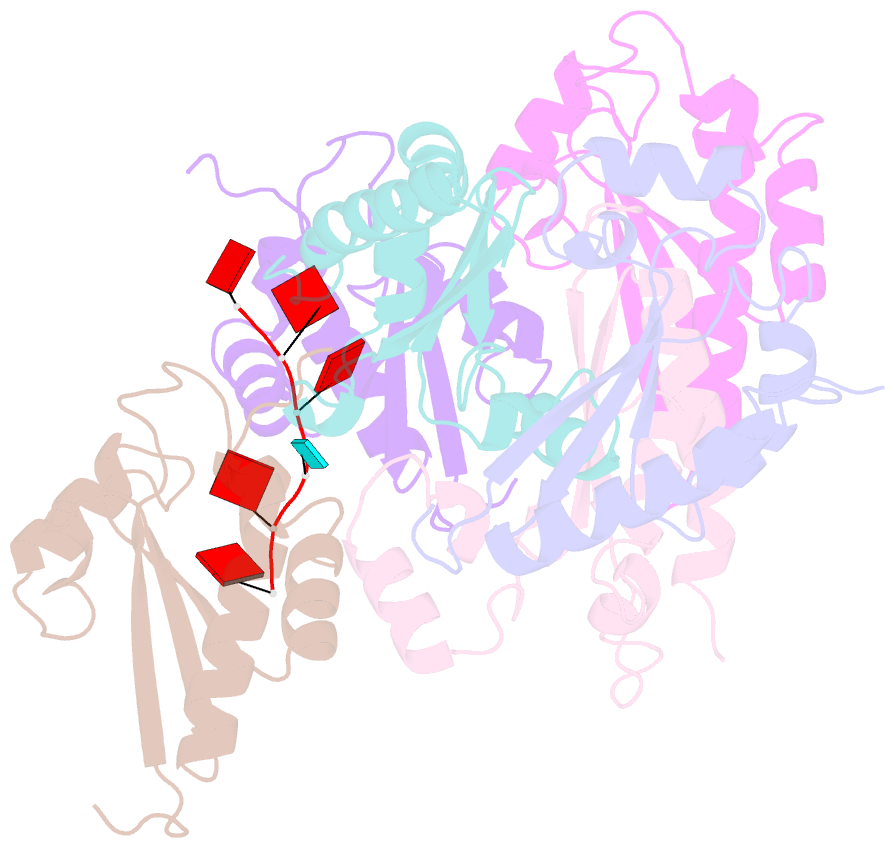
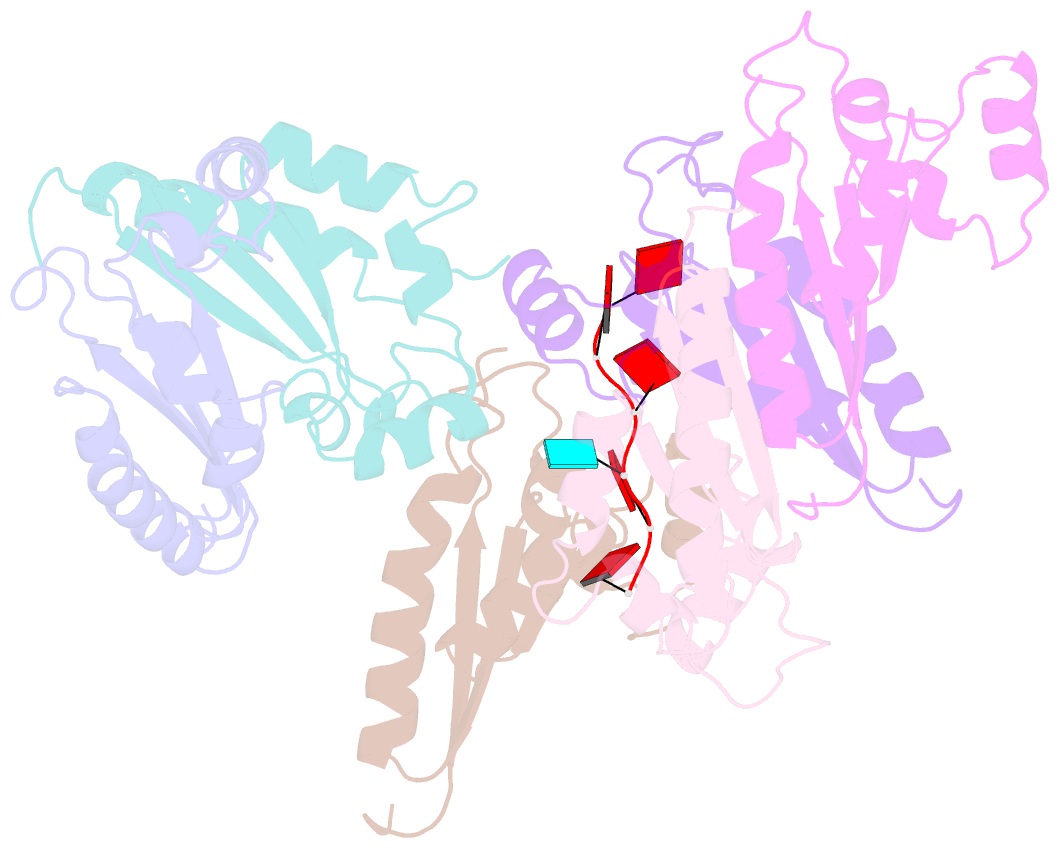
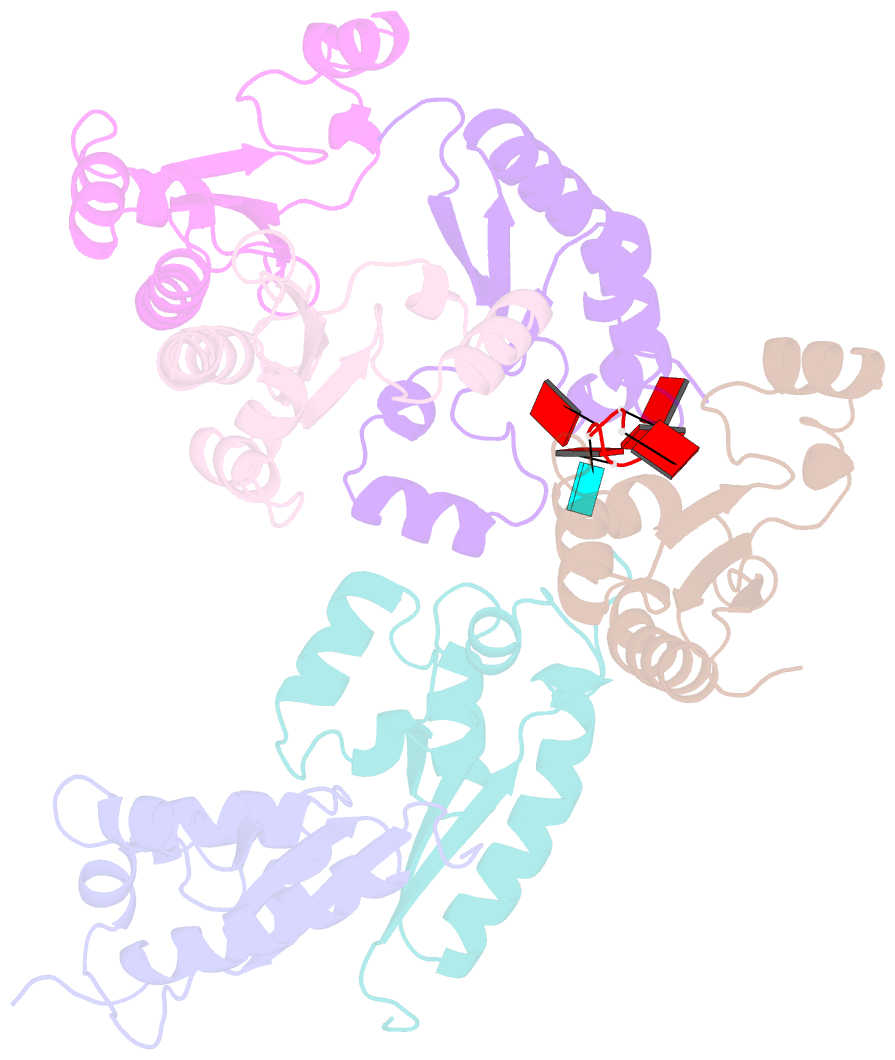
- Structure of the kh domain of t-star in complex with aaauaa RNA
- Reference
- Feracci M, Foot JN, Grellscheid SN, Danilenko M, Stehle R, Gonchar O, Kang HS, Dalgliesh C, Meyer NH, Liu Y, Lahat A, Sattler M, Eperon IC, Elliott DJ, Dominguez C (2016): "Structural basis of RNA recognition and dimerization by the STAR proteins T-STAR and Sam68." Nat Commun, 7, 10355. doi: 10.1038/ncomms10355.
- Abstract
- Sam68 and T-STAR are members of the STAR family of proteins that directly link signal transduction with post-transcriptional gene regulation. Sam68 controls the alternative splicing of many oncogenic proteins. T-STAR is a tissue-specific paralogue that regulates the alternative splicing of neuronal pre-mRNAs. STAR proteins differ from most splicing factors, in that they contain a single RNA-binding domain. Their specificity of RNA recognition is thought to arise from their property to homodimerize, but how dimerization influences their function remains unknown. Here, we establish at atomic resolution how T-STAR and Sam68 bind to RNA, revealing an unexpected mode of dimerization different from other members of the STAR family. We further demonstrate that this unique dimerization interface is crucial for their biological activity in splicing regulation, and suggest that the increased RNA affinity through dimer formation is a crucial parameter enabling these proteins to select their functional targets within the transcriptome.