Summary information and primary citation
- PDB-id
- 5elx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (1.81 Å)
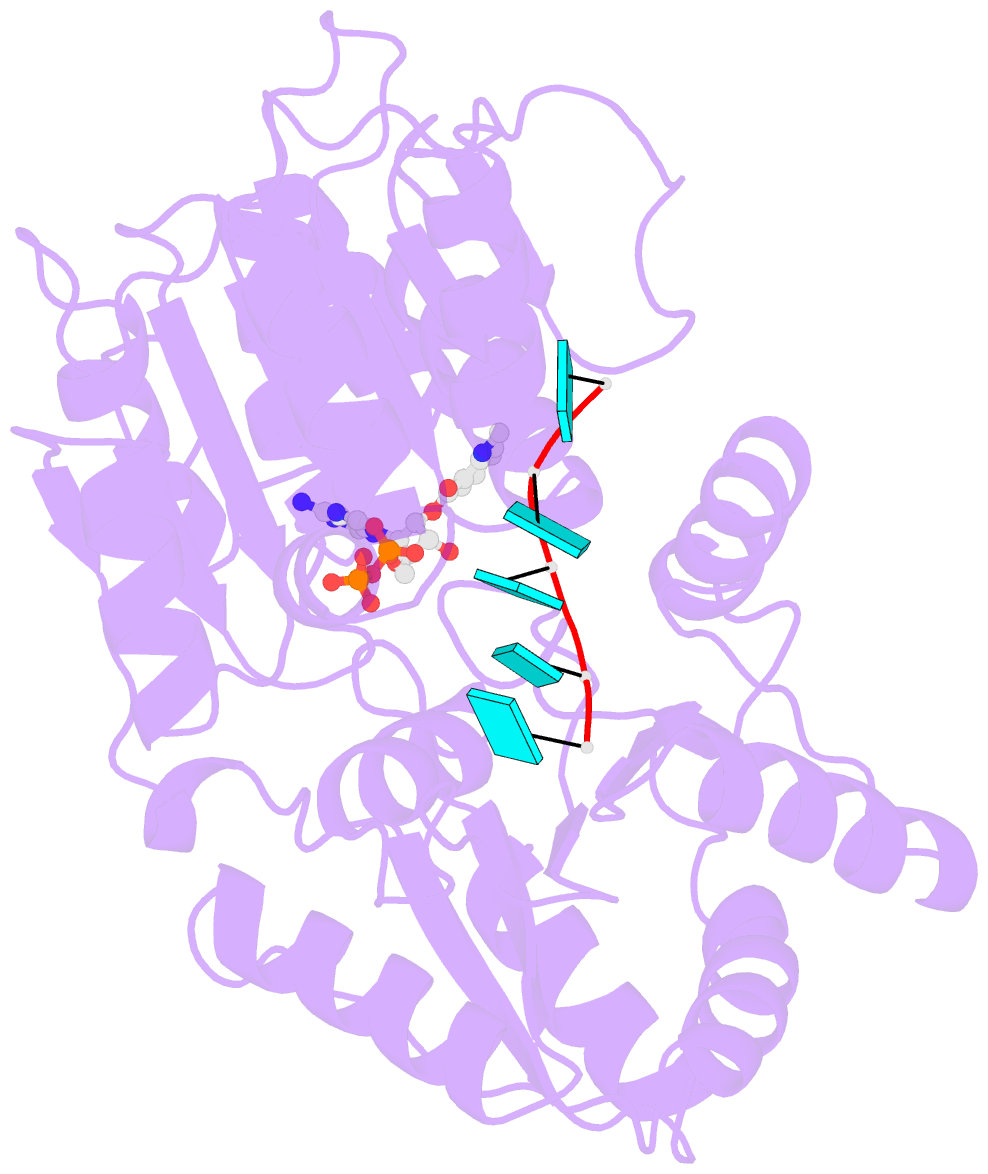
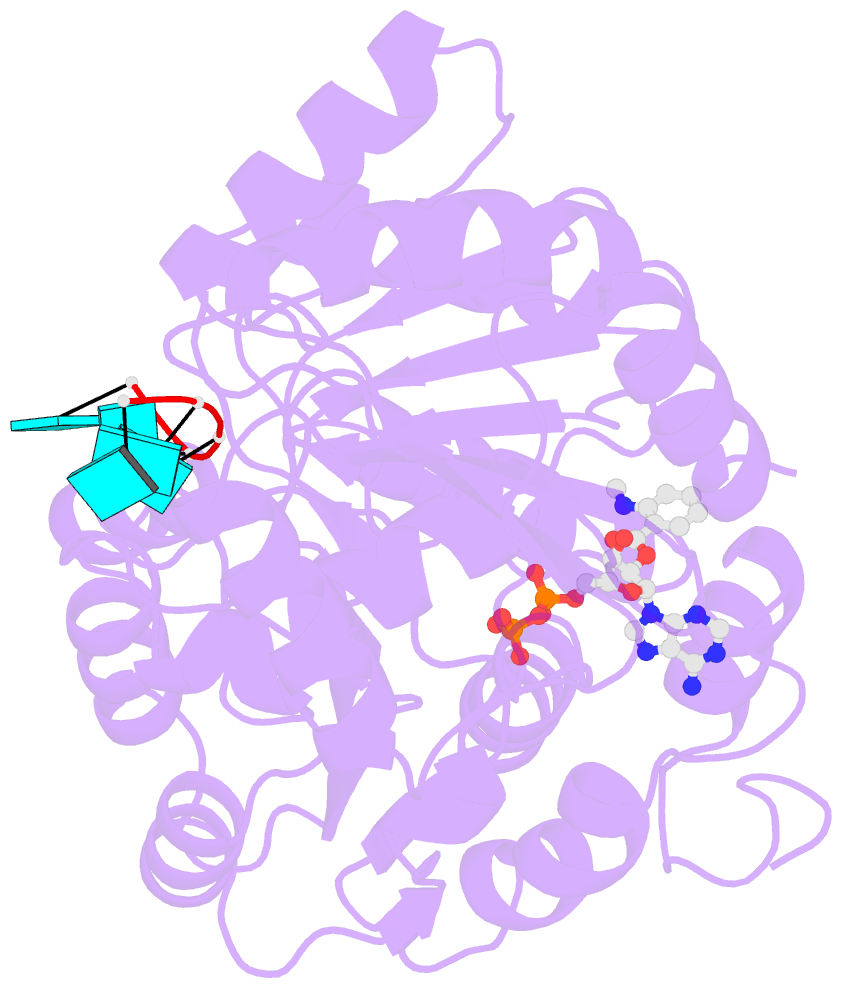
- Summary
- S. cerevisiae dbp5 bound to RNA and mant-adp bef3
- Reference
- Wong EV, Cao W, Voros J, Merchant M, Modis Y, Hackney DD, Montpetit B, De La Cruz EM (2016): "Pi Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5)." J.Mol.Biol., 428, 492-508. doi: 10.1016/j.jmb.2015.12.018.
- Abstract
- mRNA export from the nucleus depends on the ATPase activity of the DEAD-box protein Dbp5/DDX19. Although Dbp5 has measurable ATPase activity alone, several regulatory factors (e.g., RNA, nucleoporin proteins, and the endogenous small molecule InsP6) modulate catalytic activity in vitro and in vivo to facilitate mRNA export. An analysis of the intrinsic and regulator-activated Dbp5 ATPase cycle is necessary to define how these factors control Dbp5 and mRNA export. Here, we report a kinetic and equilibrium analysis of the Saccharomyces cerevisiae Dbp5 ATPase cycle, including the influence of RNA on Dbp5 activity. These data show that ATP binds Dbp5 weakly in rapid equilibrium with a binding affinity (KT~4 mM) comparable to the KM for steady-state cycling, while ADP binds an order of magnitude more tightly (KD~0.4 mM). The overall intrinsic steady-state cycling rate constant (kcat) is limited by slow, near-irreversible ATP hydrolysis and even slower subsequent phosphate release. RNA increases kcat and rate-limiting Pi release 20-fold, although Pi release continues to limit steady-state cycling in the presence of RNA, in conjunction with RNA binding. Together, this work identifies RNA binding and Pi release as important biochemical transitions within the Dbp5 ATPase cycle and provides a framework for investigating the means by which Dbp5 and mRNA export is modulated by regulatory factors.