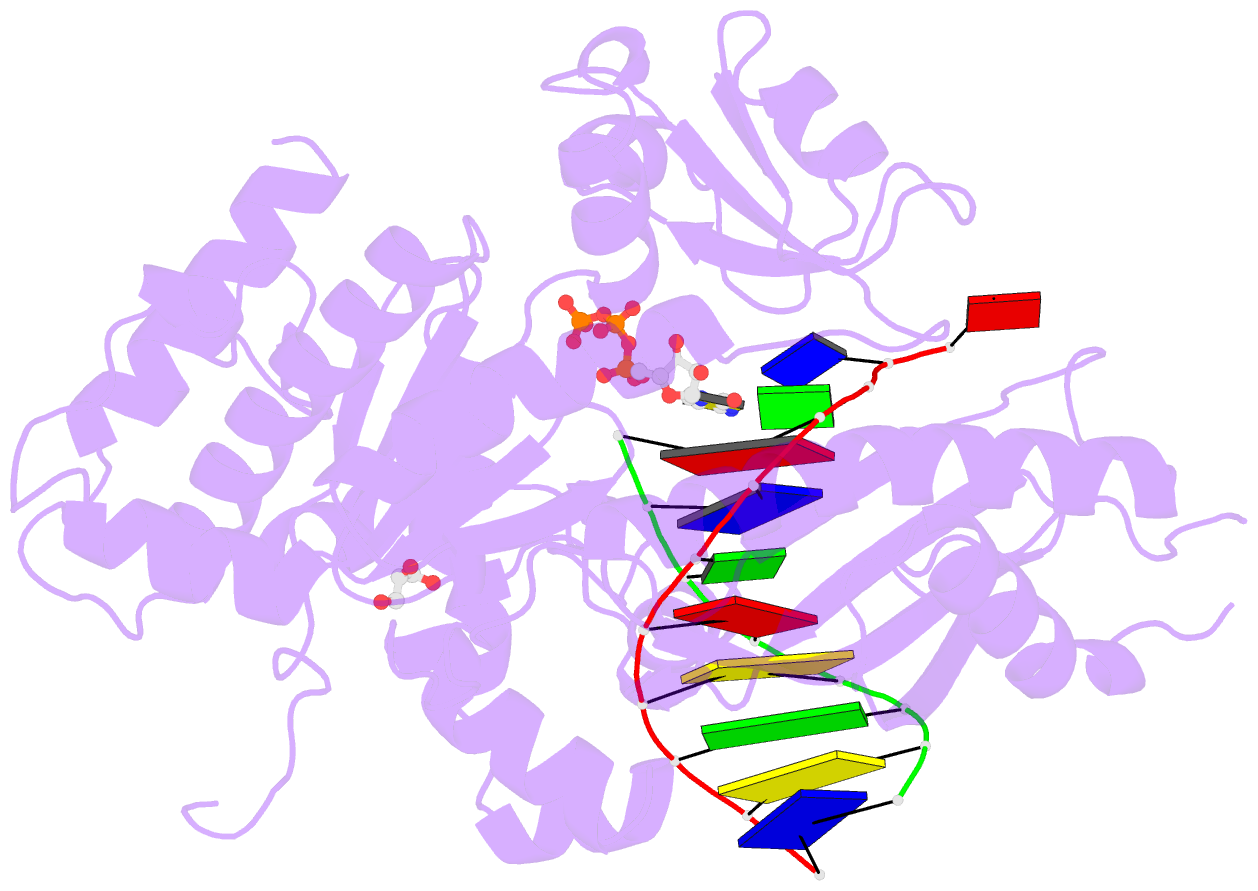
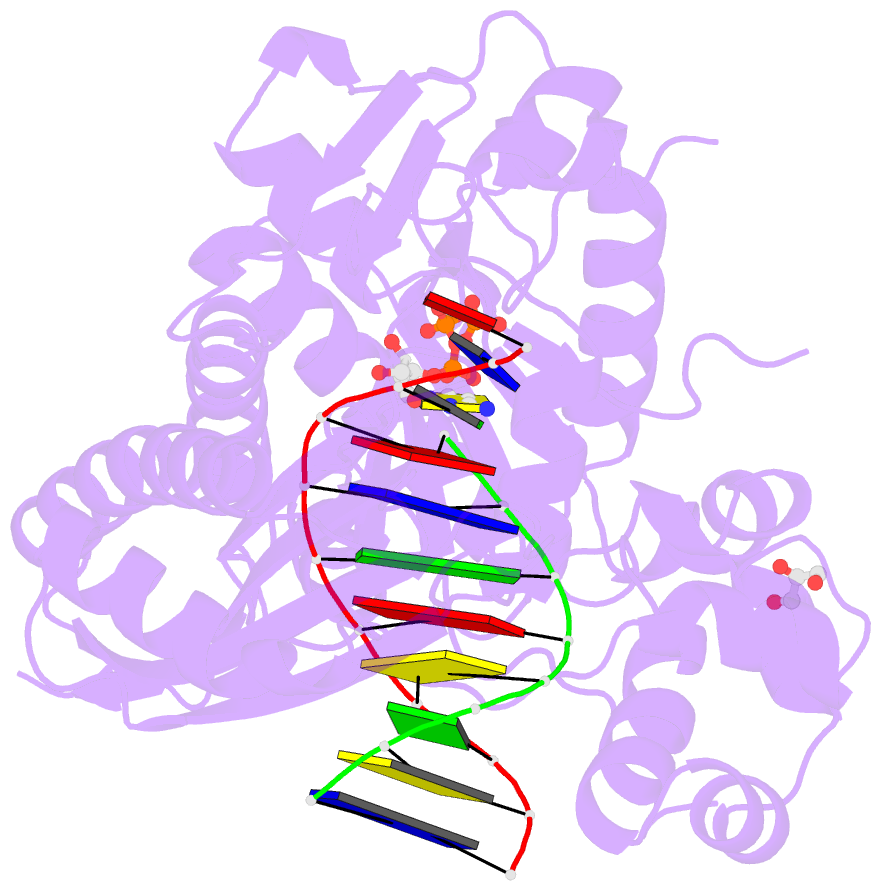
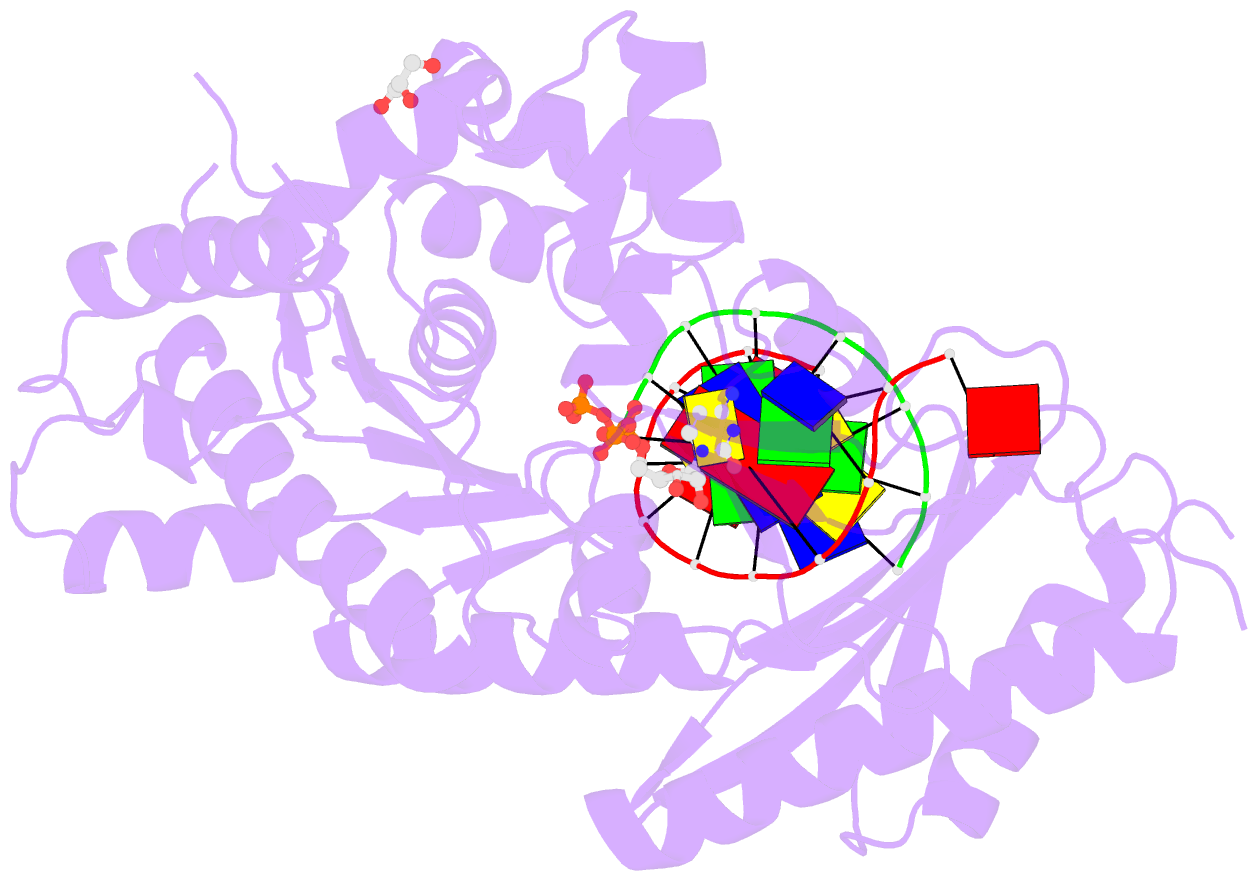
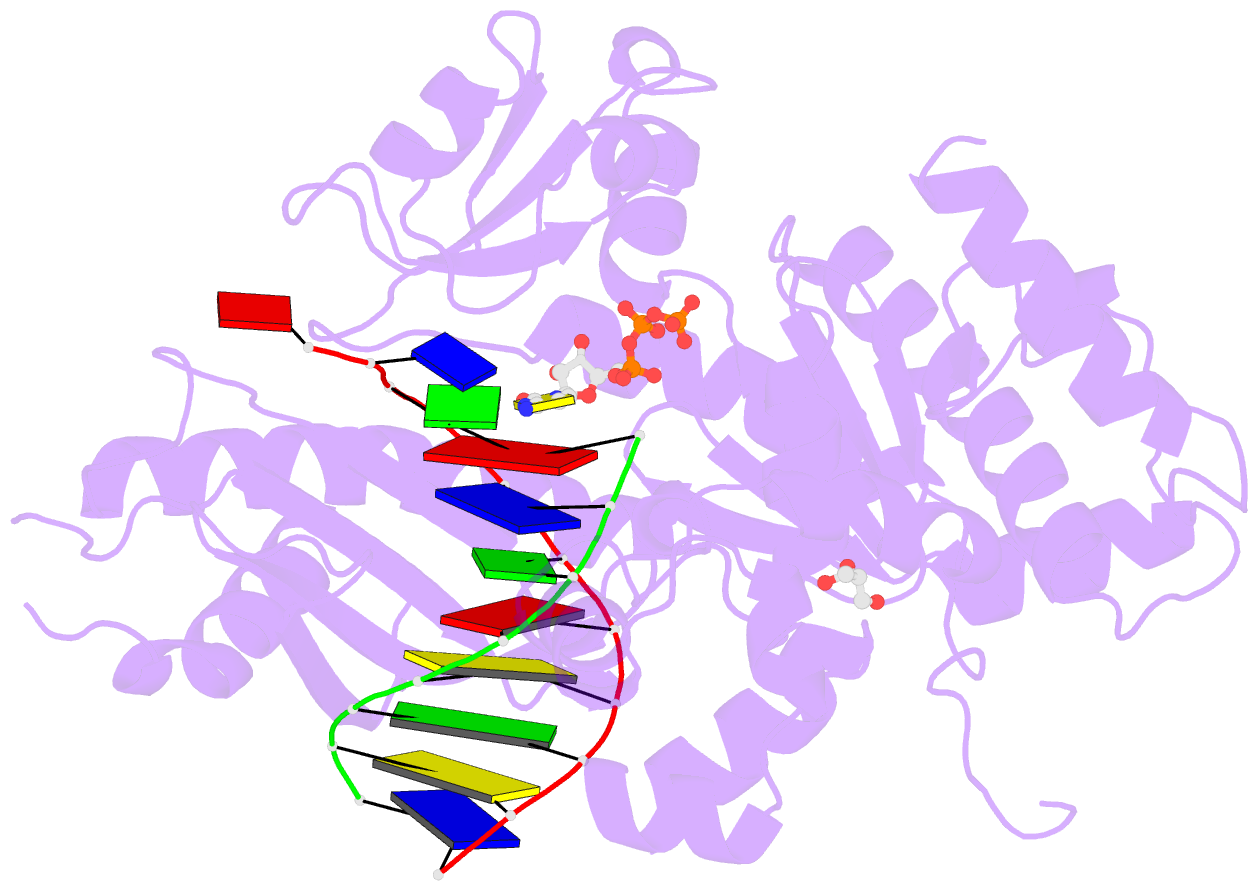
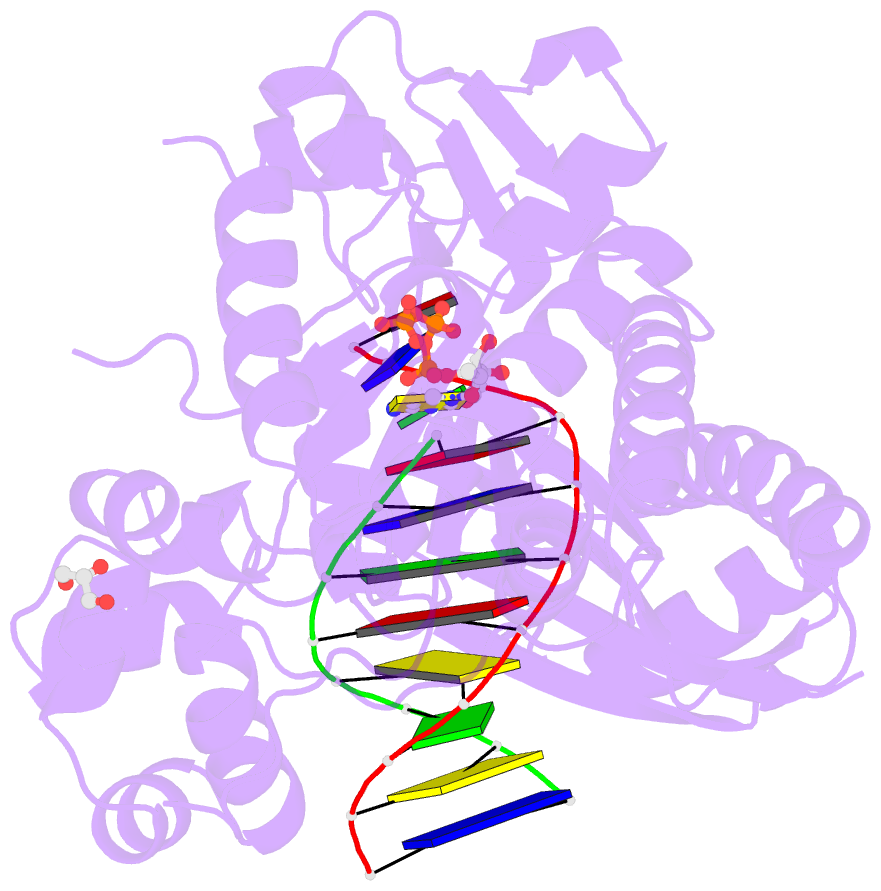
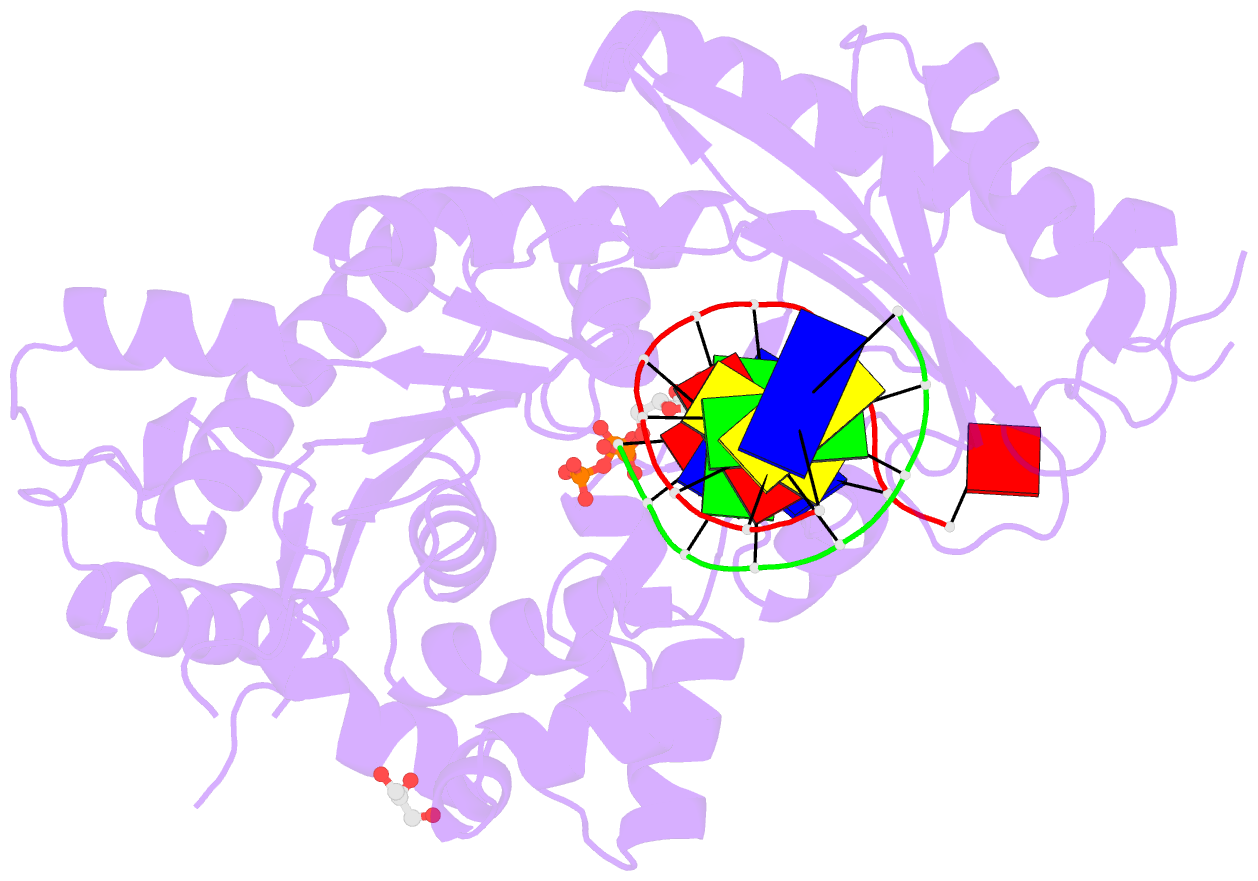
Summary information and primary citation
- PDB-id
- 5ewf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.782 Å)
- Summary
- Ternary complex of human DNA polymerase eta inserting rctp opposite an 8-oxodeoxyguanosine lesion
- Reference
- Su Y, Egli M, Guengerich FP (2016): "Mechanism of Ribonucleotide Incorporation by Human DNA Polymerase eta." J.Biol.Chem., 291, 3747-3756. doi: 10.1074/jbc.M115.706226.
- Abstract
- Ribonucleotides and 2'-deoxyribonucleotides are the basic units for RNA and DNA, respectively, and the only difference is the extra 2'-OH group on the ribonucleotide sugar. Cellular rNTP concentrations are much higher than those of dNTP. When copying DNA, DNA polymerases not only select the base of the incoming dNTP to form a Watson-Crick pair with the template base but also distinguish the sugar moiety. Some DNA polymerases use a steric gate residue to prevent rNTP incorporation by creating a clash with the 2'-OH group. Y-family human DNA polymerase η (hpol η) is of interest because of its spacious active site (especially in the major groove) and tolerance of DNA lesions. Here, we show that hpol η maintains base selectivity when incorporating rNTPs opposite undamaged DNA and the DNA lesions 7,8-dihydro-8-oxo-2'-deoxyguanosine and cyclobutane pyrimidine dimer but with rates that are 10(3)-fold lower than for inserting the corresponding dNTPs. X-ray crystal structures show that the hpol η scaffolds the incoming rNTP to pair with the template base (dG) or 7,8-dihydro-8-oxo-2'-deoxyguanosine with a significant propeller twist. As a result, the 2'-OH group avoids a clash with the steric gate, Phe-18, but the distance between primer end and Pα of the incoming rNTP increases by 1 Å, elevating the energy barrier and slowing polymerization compared with dNTP. In addition, Tyr-92 was identified as a second line of defense to maintain the position of Phe-18. This is the first crystal structure of a DNA polymerase with an incoming rNTP opposite a DNA lesion.