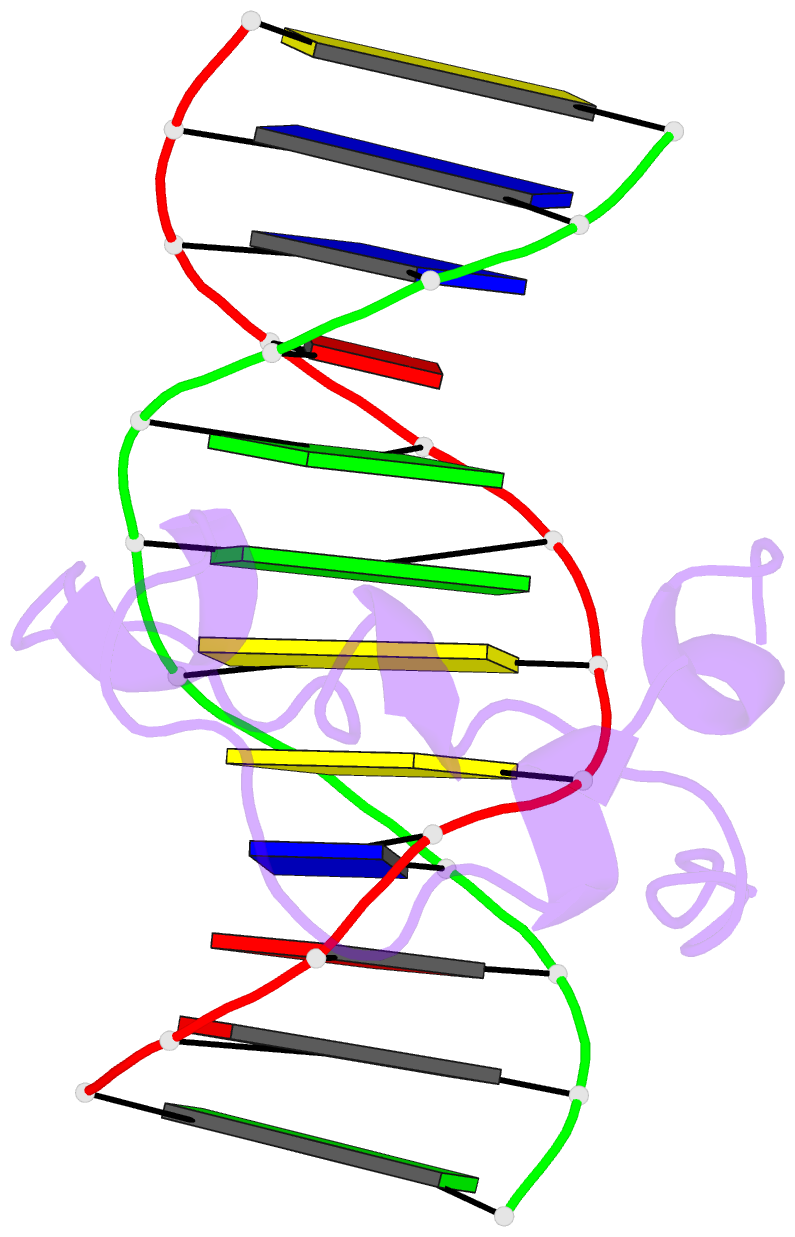
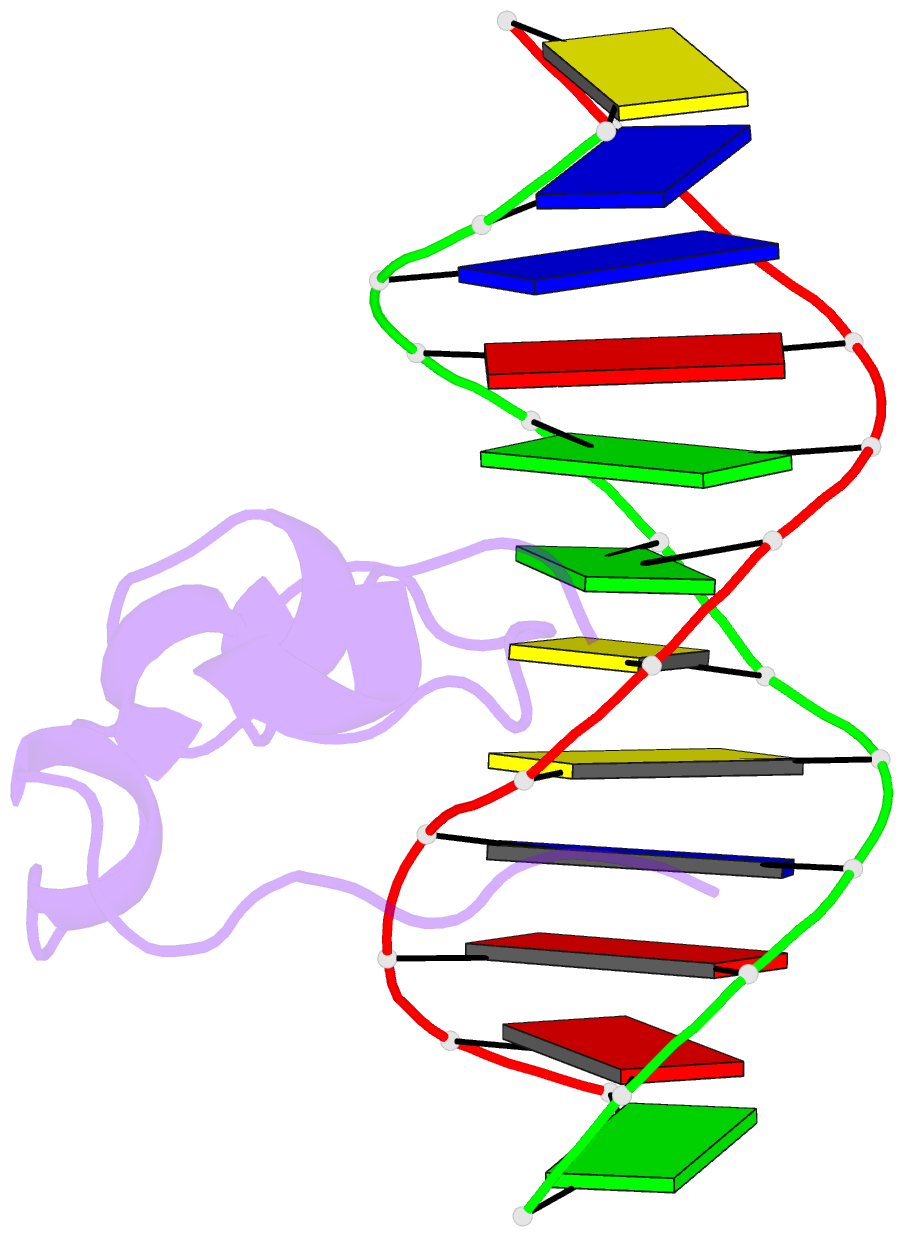
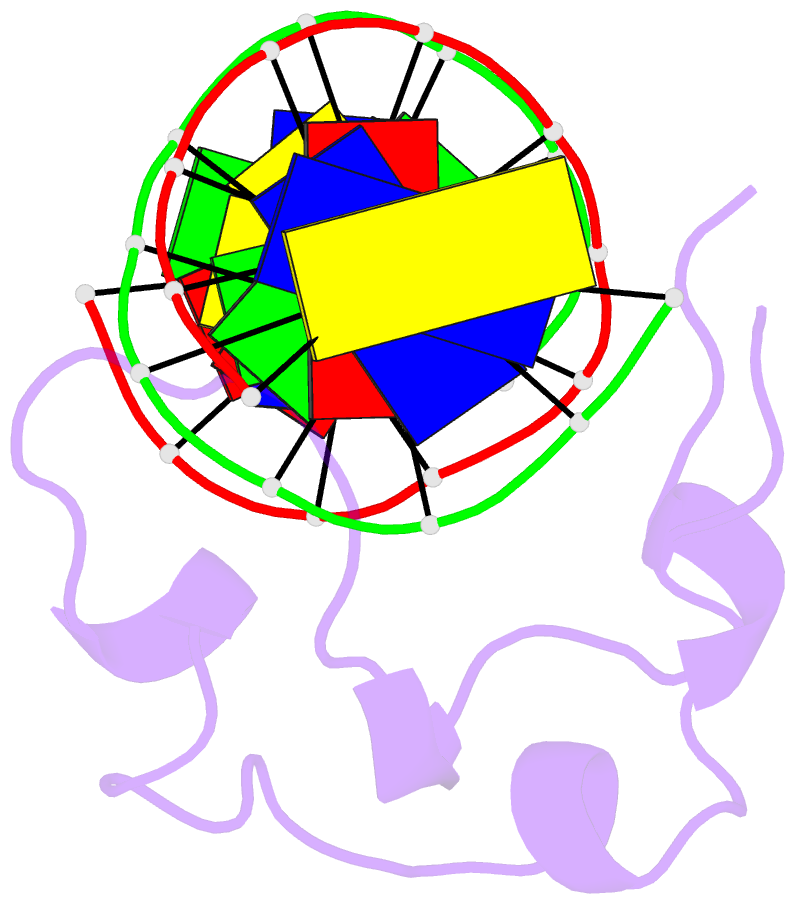
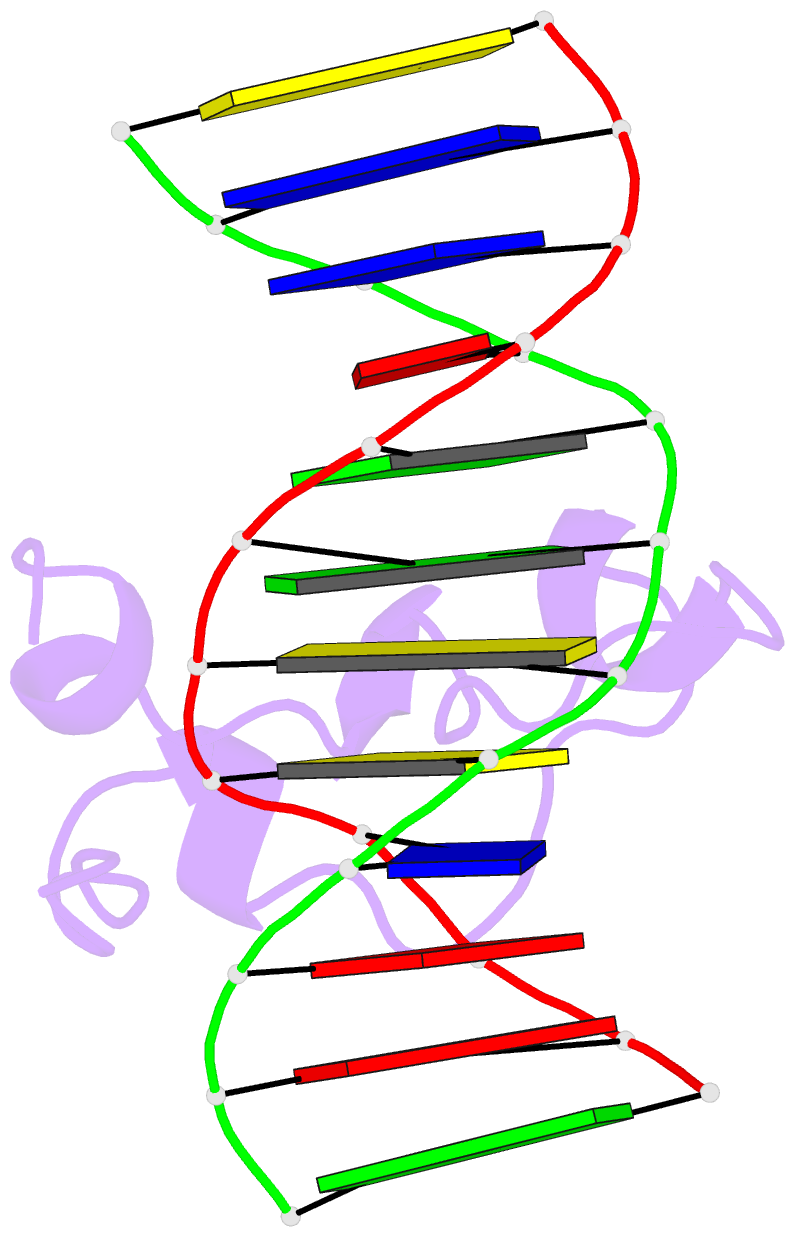
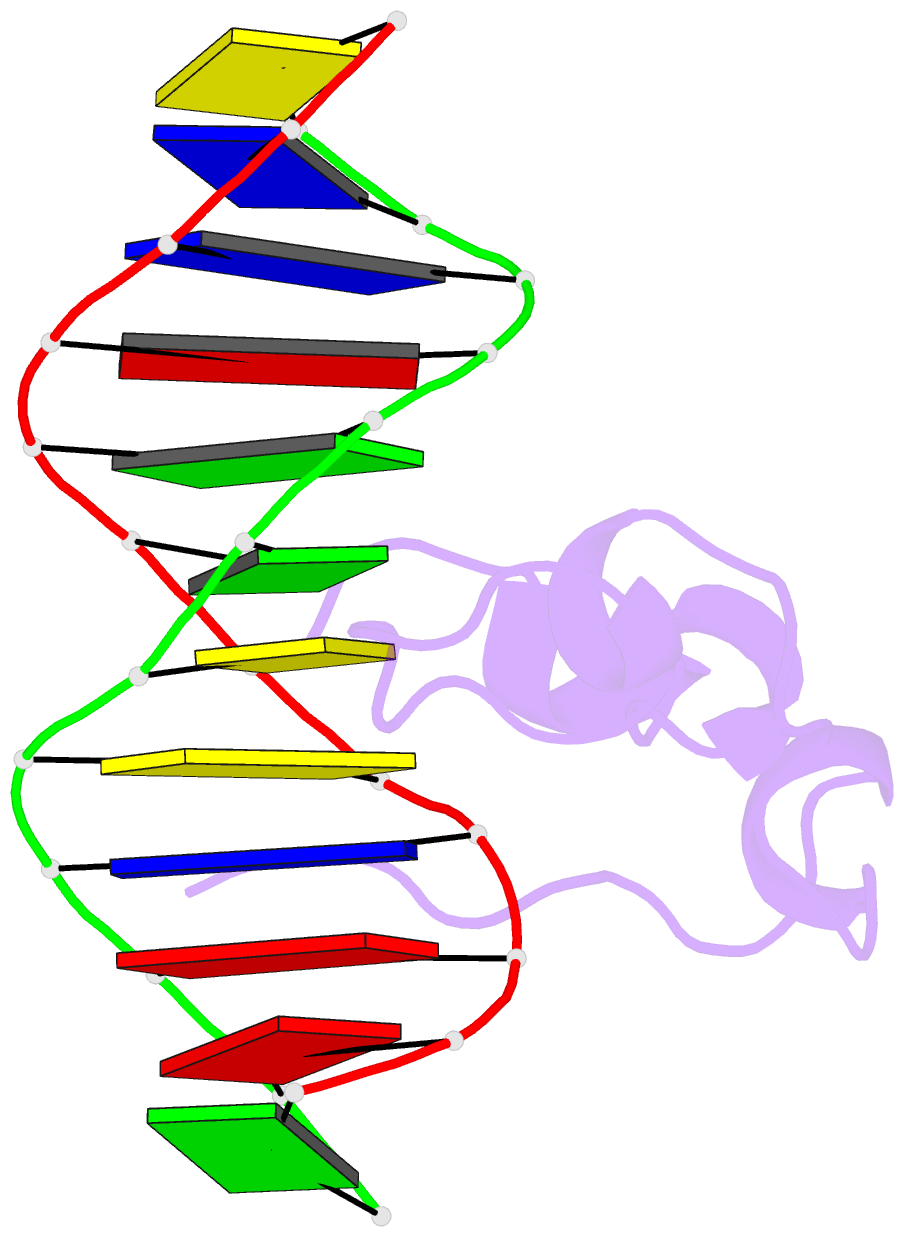
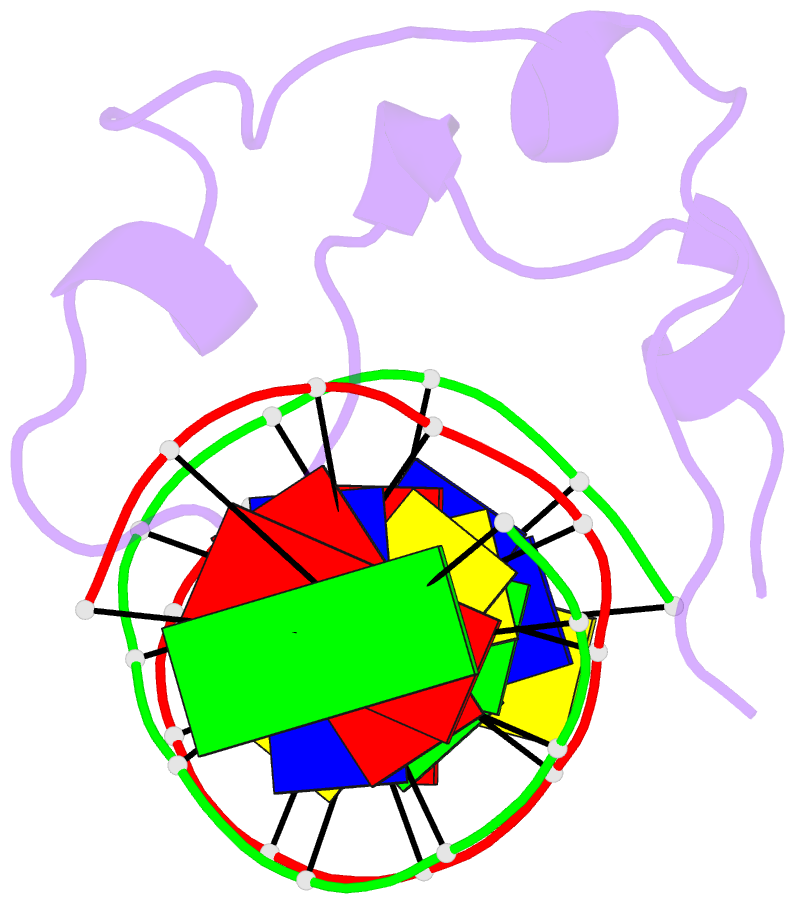
Summary information and primary citation
- PDB-id
-
5exh;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
- oxidoreductase-DNA
- Method
- X-ray (1.3 Å)
- Summary
- Crystal structure of mtet3-cxxc domain in complex with
5-carboxylcytosine DNA at 1.3 angstroms resolution.
- Reference
-
Jin SG, Zhang ZM, Dunwell TL, Harter MR, Wu X, Johnson J,
Li Z, Liu J, Szabo PE, Lu Q, Xu GL, Song J, Pfeifer GP
(2016): "Tet3 Reads
5-Carboxylcytosine through Its CXXC Domain and Is a
Potential Guardian against Neurodegeneration."
Cell Rep, 14, 493-505. doi:
10.1016/j.celrep.2015.12.044.
- Abstract
- We report that the mammalian 5-methylcytosine (5mC)
oxidase Tet3 exists as three major isoforms and
characterized the full-length isoform containing an
N-terminal CXXC domain (Tet3FL). This CXXC domain binds to
unmethylated CpGs, but, unexpectedly, its highest affinity
is toward 5-carboxylcytosine (5caC). We determined the
crystal structure of the CXXC domain-5caC-DNA complex,
revealing the structural basis of the binding specificity
of this domain as a reader of CcaCG sequences. Mapping of
Tet3FL in neuronal cells shows that Tet3FL is localized
precisely at the transcription start sites (TSSs) of genes
involved in lysosome function, mRNA processing, and key
genes of the base excision repair pathway. Therefore,
Tet3FL may function as a regulator of 5caC removal by base
excision repair. Active removal of accumulating 5mC from
the TSSs of genes coding for lysosomal proteins by Tet3FL
in postmitotic neurons of the brain may be important for
preventing neurodegenerative diseases.