Summary information and primary citation
- PDB-id
- 5fkw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- cryo-EM (7.3 Å)
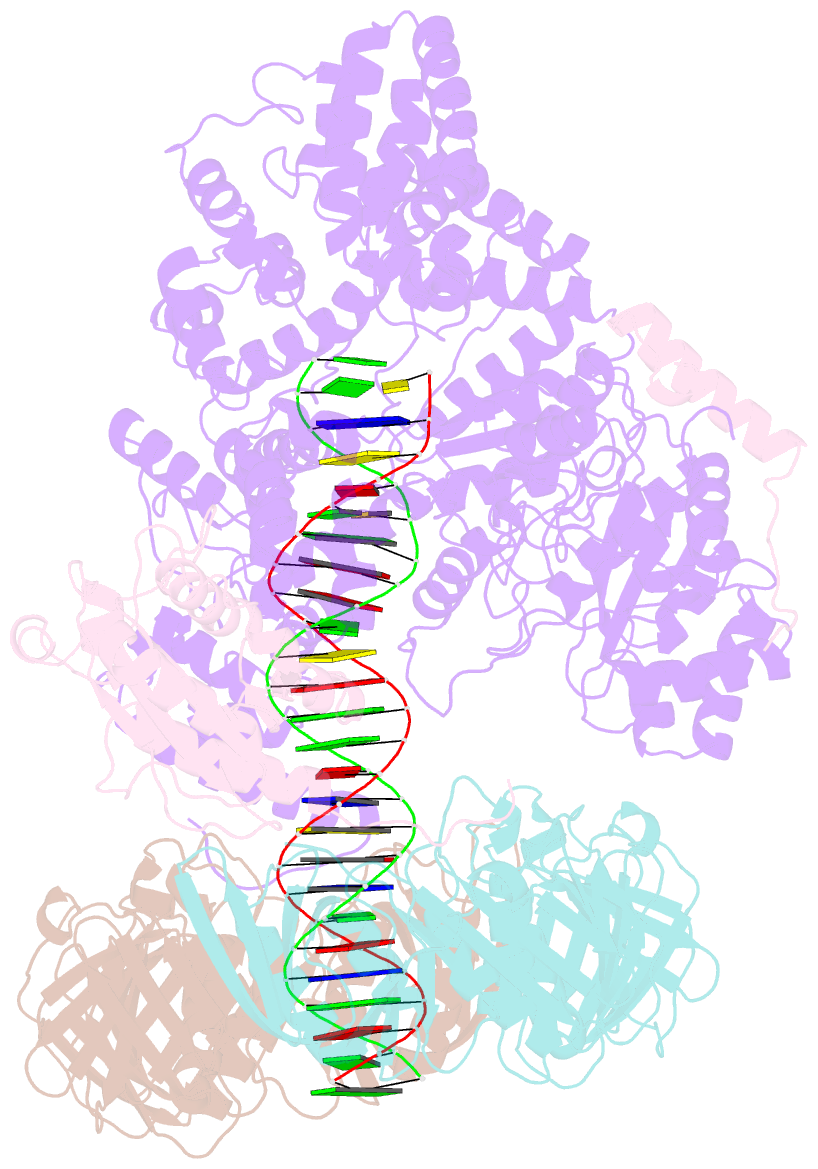
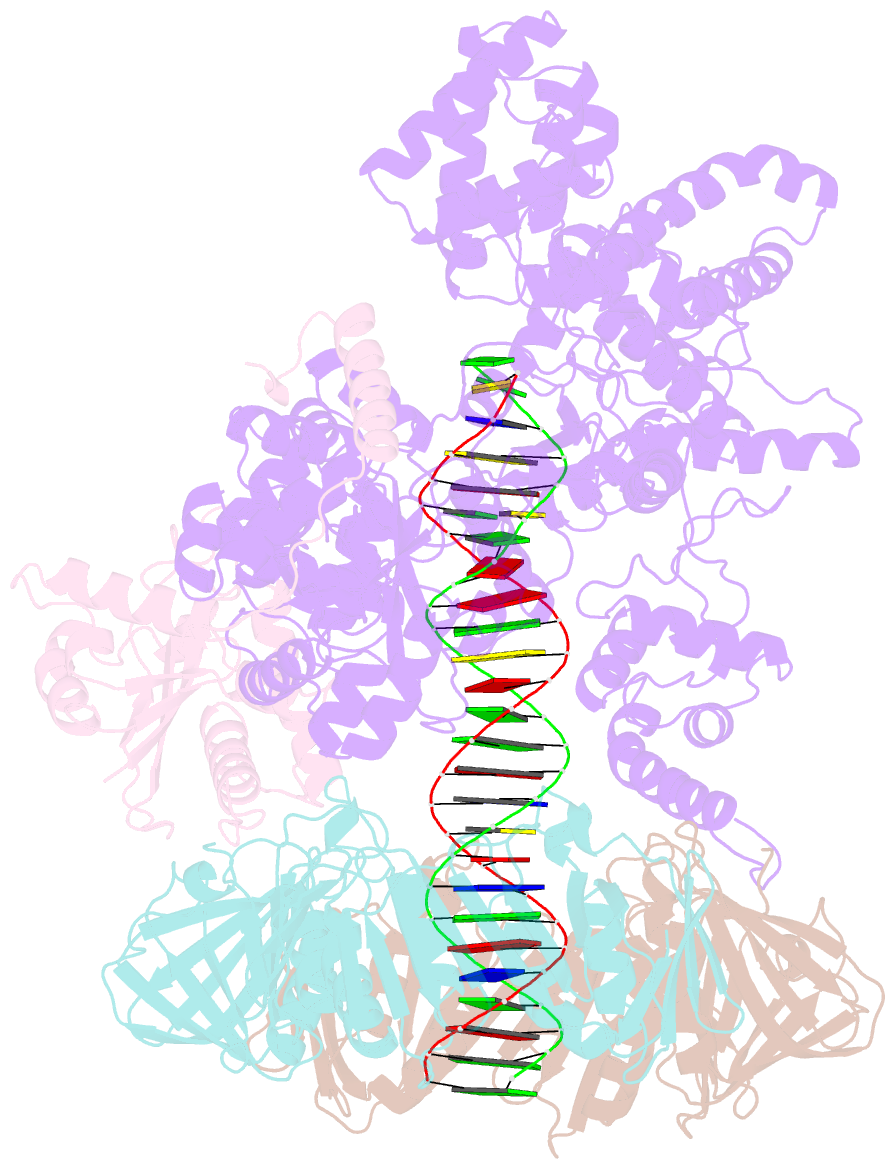
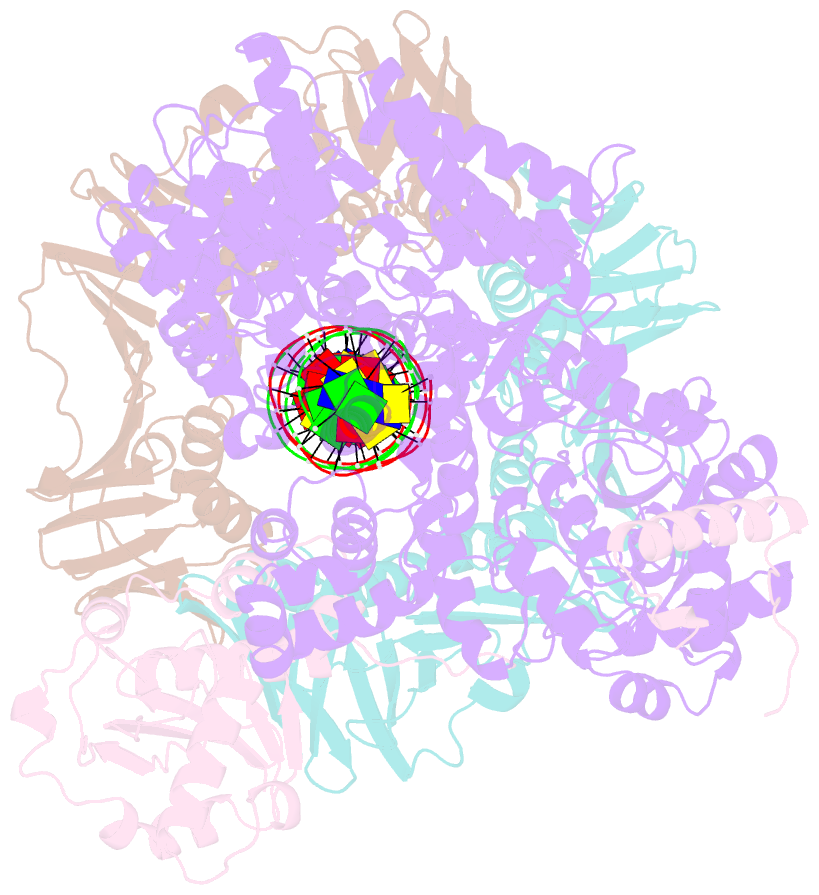
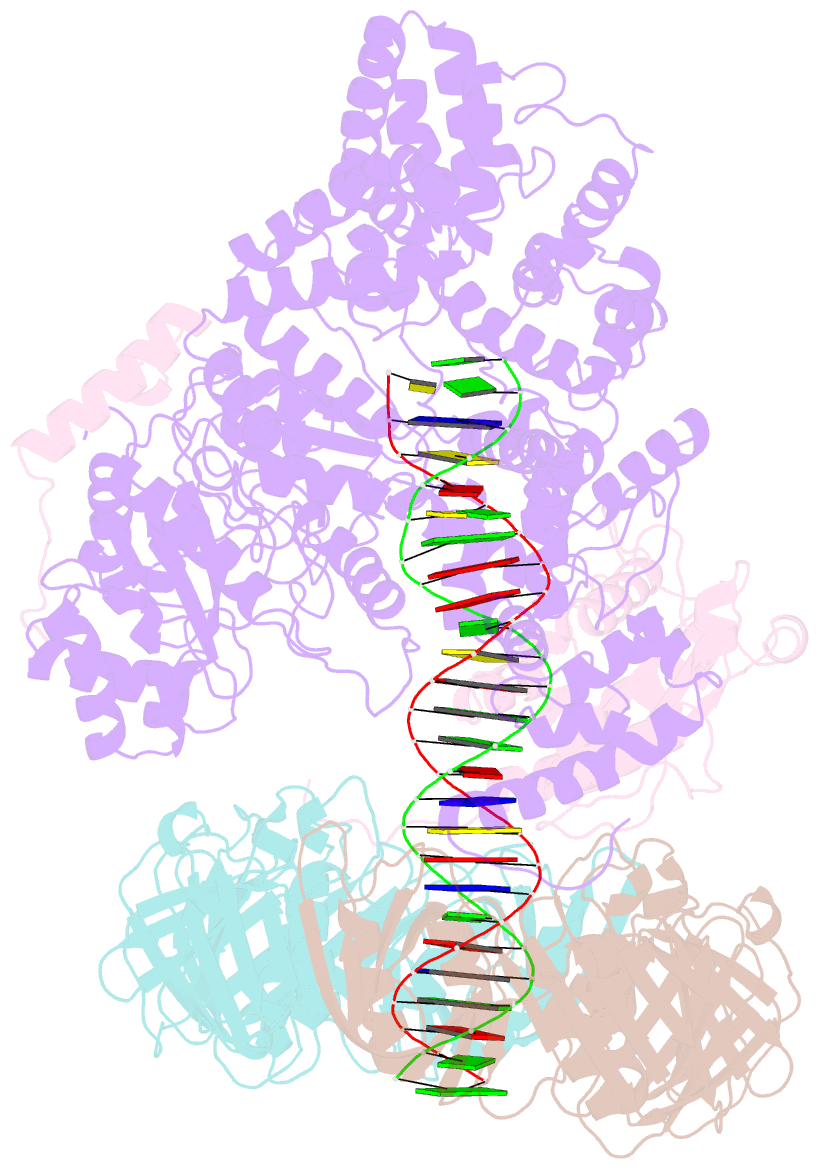
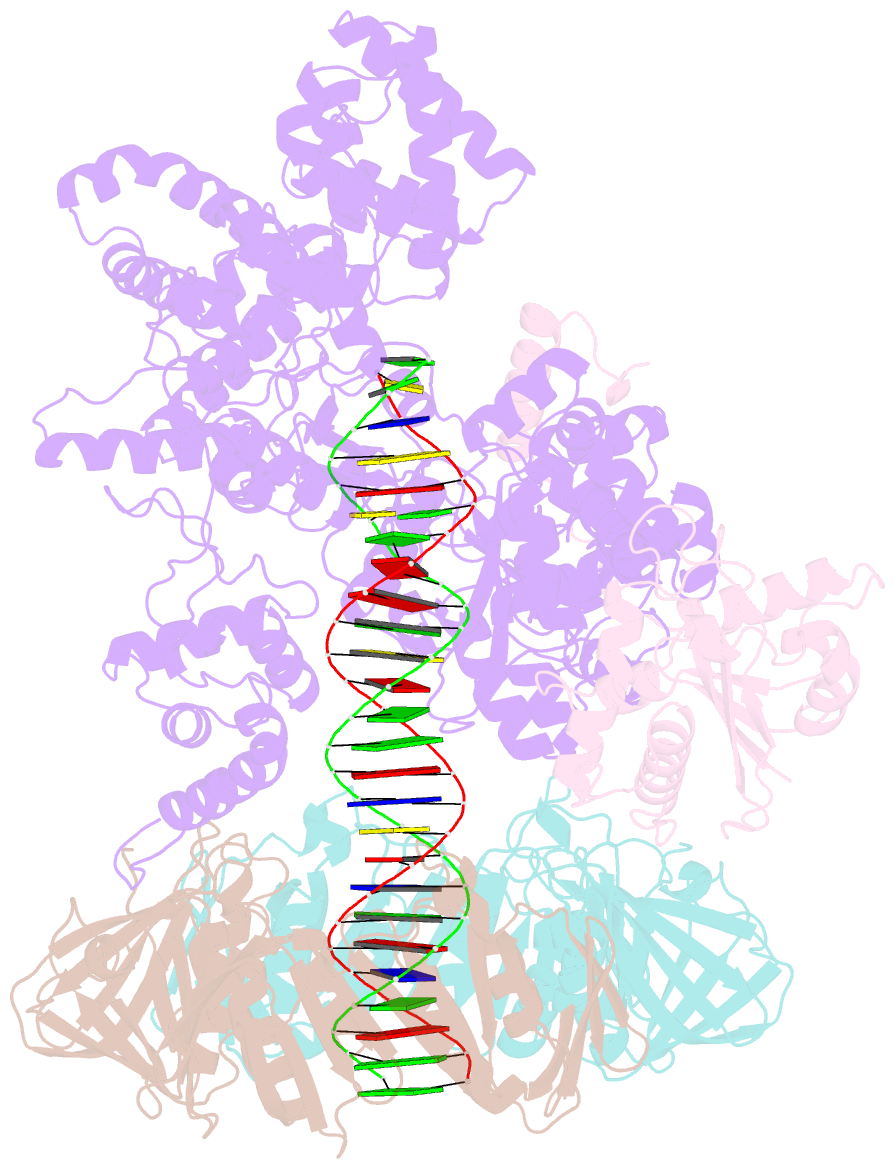
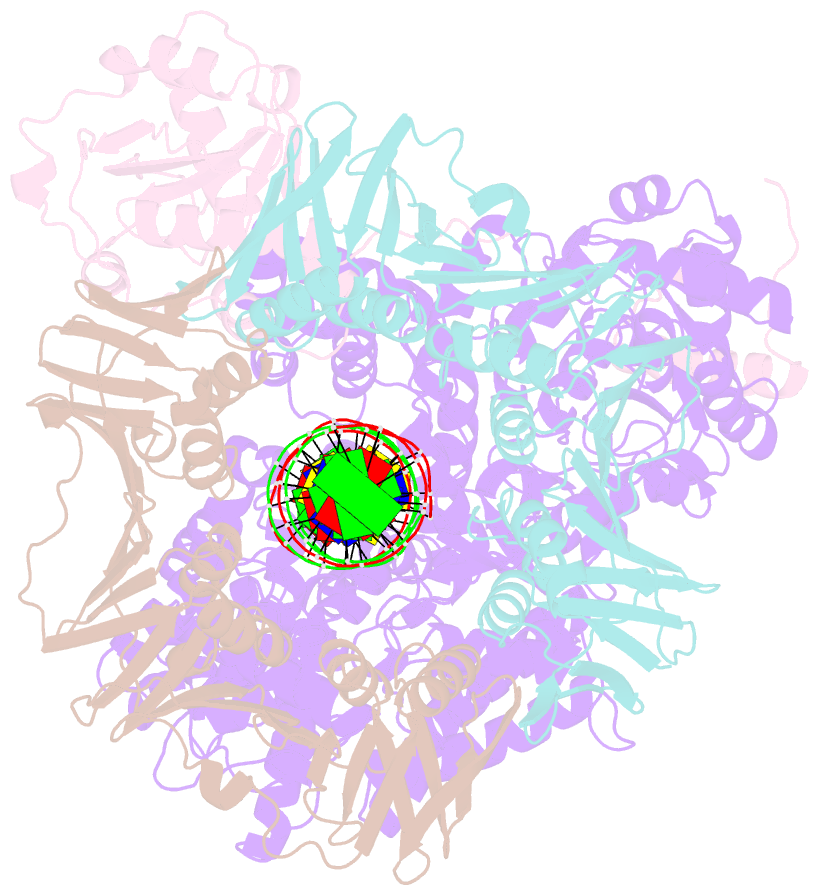
- Summary
- cryo-EM structure of the e. coli replicative DNA polymerase complex bound to DNA (DNA polymerase iii alpha, beta, epsilon)
- Reference
- Fernandez-Leiro R, Conrad J, Scheres SH, Lamers MH (2015): "cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau." Elife, 4. doi: 10.7554/eLife.11134.
- Abstract
- The replicative DNA polymerase PolIIIα from Escherichia coli is a uniquely fast and processive enzyme. For its activity it relies on the DNA sliding clamp β, the proofreading exonuclease ε and the C-terminal domain of the clamp loader subunit τ. Due to the dynamic nature of the four-protein complex it has long been refractory to structural characterization. Here we present the 8 Å resolution cryo-electron microscopy structures of DNA-bound and DNA-free states of the PolIII-clamp-exonuclease-τc complex. The structures show how the polymerase is tethered to the DNA through multiple contacts with the clamp and exonuclease. A novel contact between the polymerase and clamp is made in the DNA bound state, facilitated by a large movement of the polymerase tail domain and τc. These structures provide crucial insights into the organization of the catalytic core of the replisome and form an important step towards determining the structure of the complete holoenzyme.