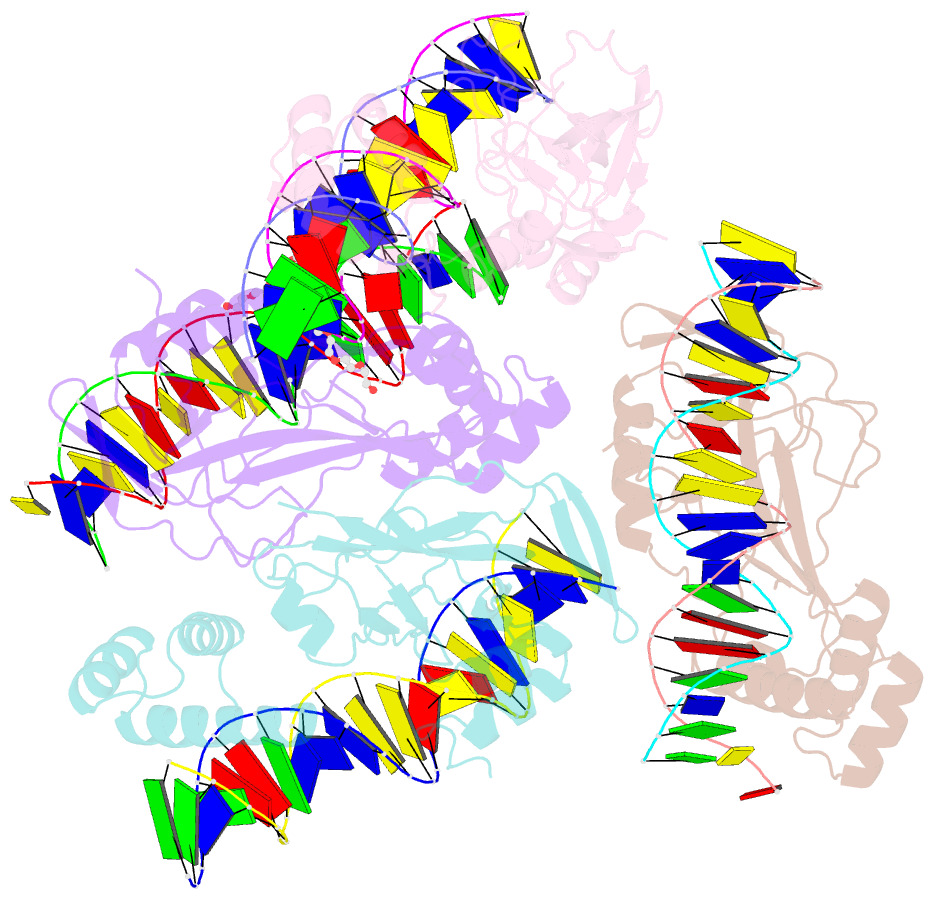
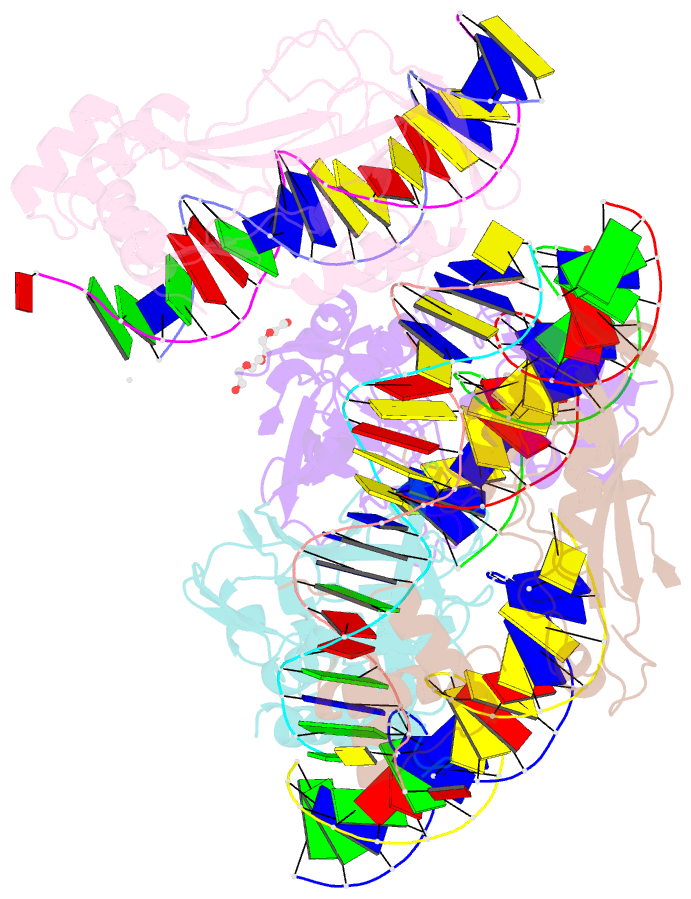
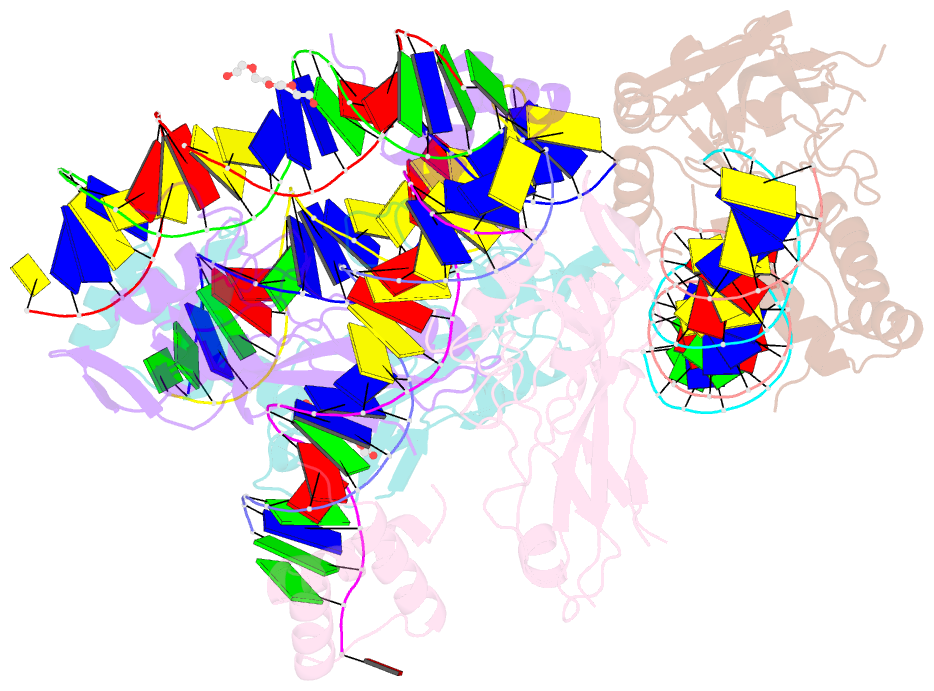
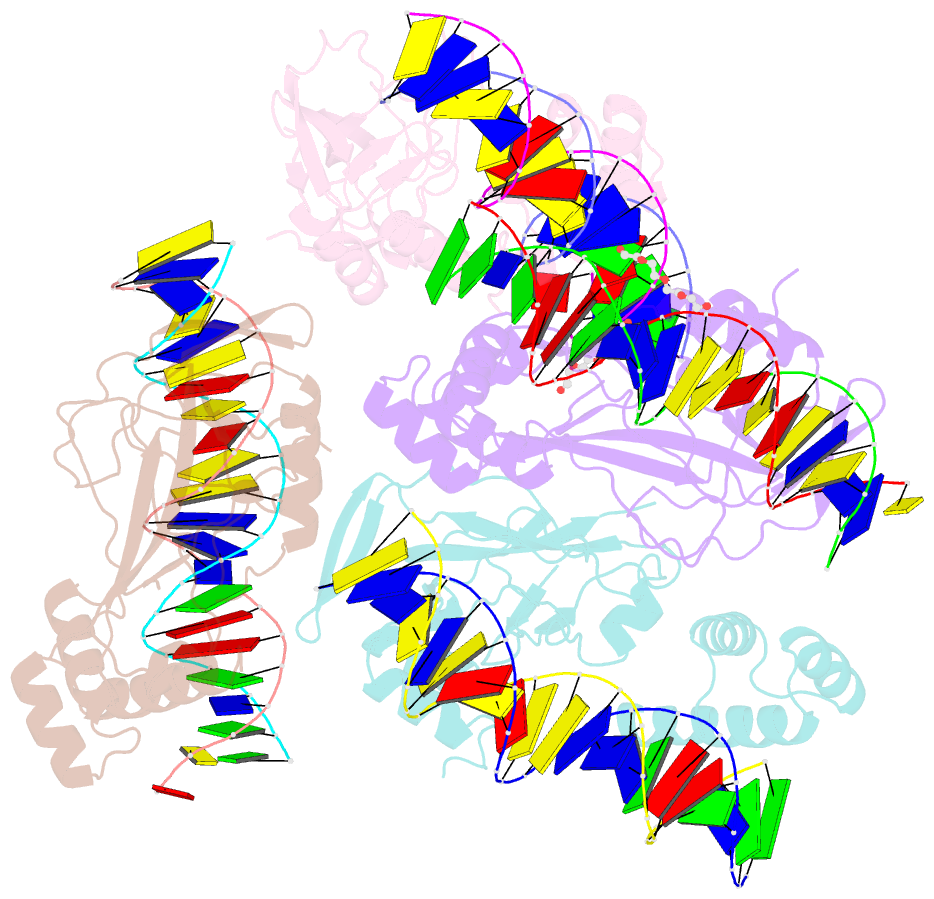
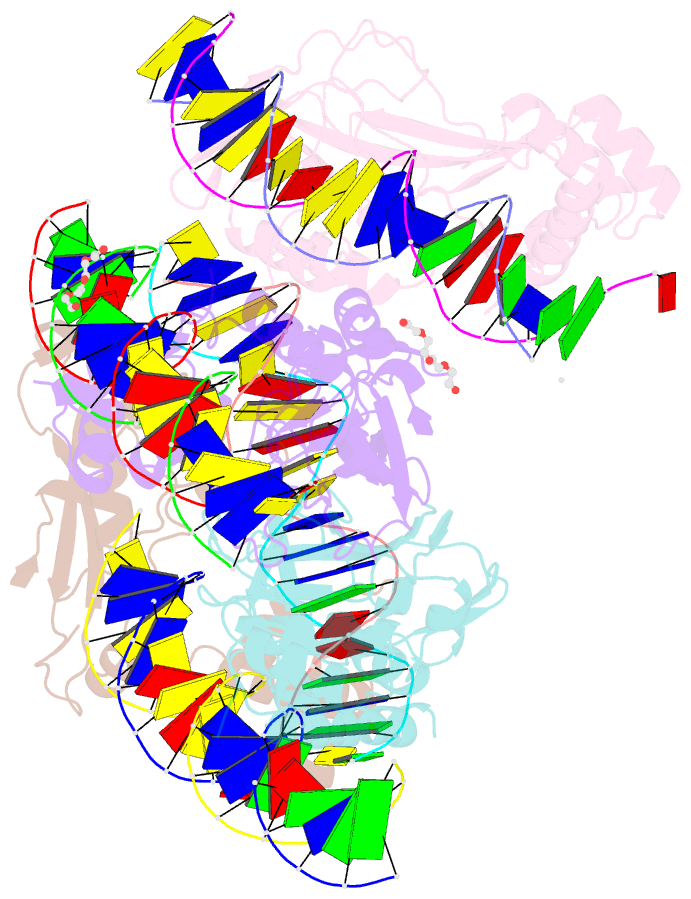
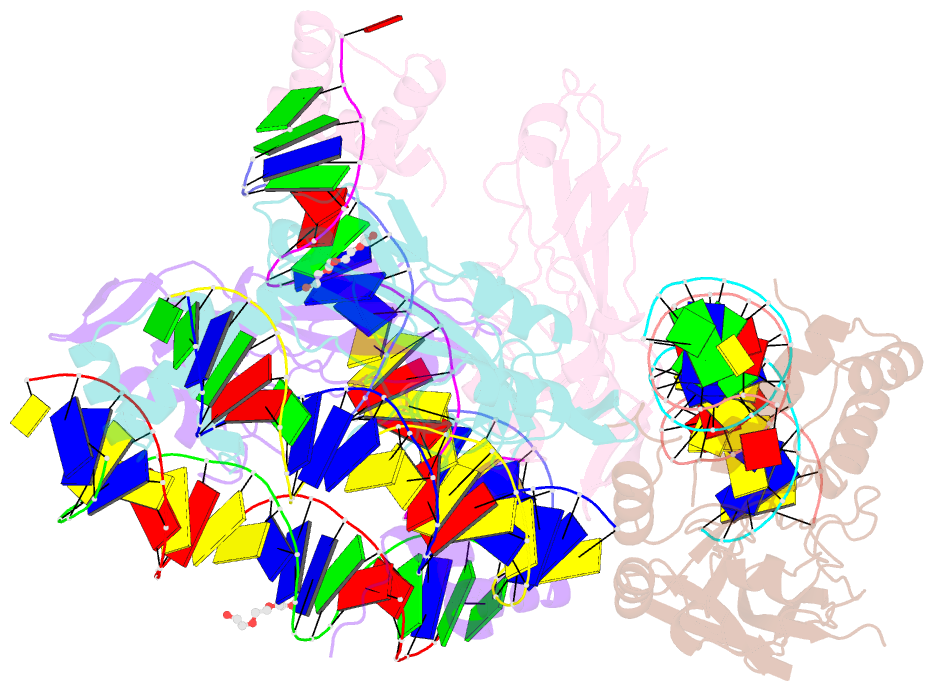
Summary information and primary citation
- PDB-id
- 5flv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (3.005 Å)
- Summary
- Crystal structure of nkx2-5 and tbx5 bound to the nppa promoter region
- Reference
- Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MAH, He D, Small EM, Mileikovsky M, Nagy A, Holloway AK, Pollard KS, Muller CW, Bruneau BG (2016): "Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis." Cell(Cambridge,Mass.), 164, 999. doi: 10.1016/J.CELL.2016.01.004.
- Abstract
- Transcription factors (TFs) are thought to function with partners to achieve specificity and precise quantitative outputs. In the developing heart, heterotypic TF interactions, such as between the T-box TF TBX5 and the homeodomain TF NKX2-5, have been proposed as a mechanism for human congenital heart defects. We report extensive and complex interdependent genomic occupancy of TBX5, NKX2-5, and the zinc finger TF GATA4 coordinately controlling cardiac gene expression, differentiation, and morphogenesis. Interdependent binding serves not only to co-regulate gene expression but also to prevent TFs from distributing to ectopic loci and activate lineage-inappropriate genes. We define preferential motif arrangements for TBX5 and NKX2-5 cooperative binding sites, supported at the atomic level by their co-crystal structure bound to DNA, revealing a direct interaction between the two factors and induced DNA bending. Complex interdependent binding mechanisms reveal tightly regulated TF genomic distribution and define a combinatorial logic for heterotypic TF regulation of differentiation.