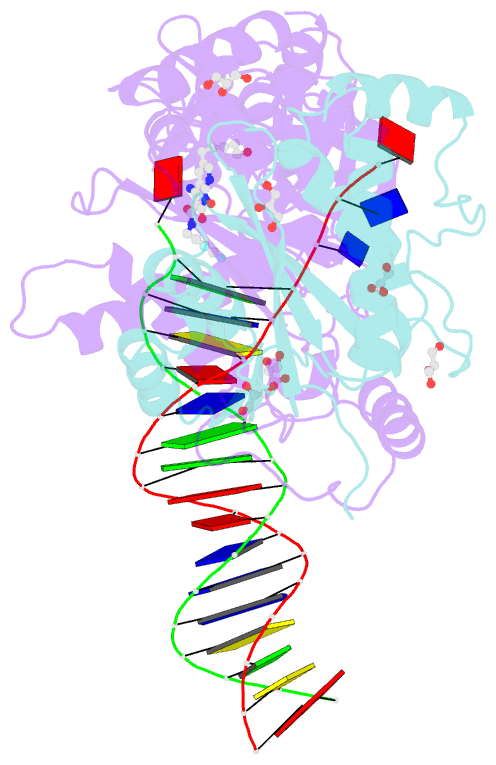
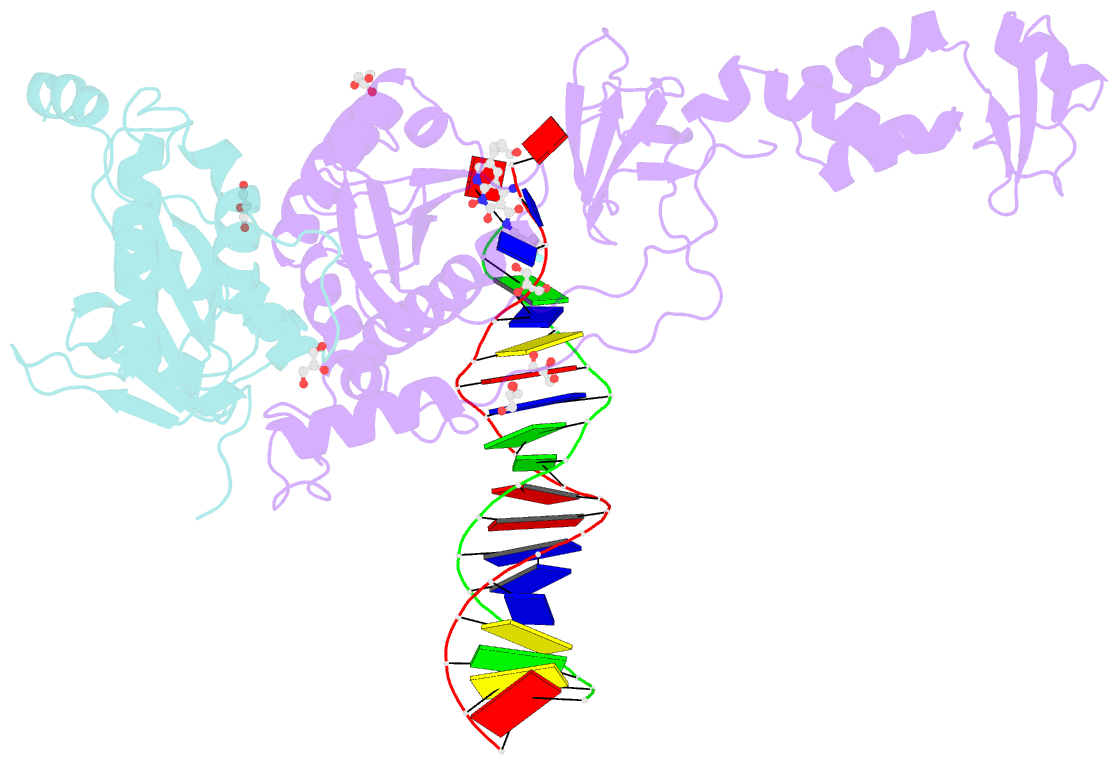
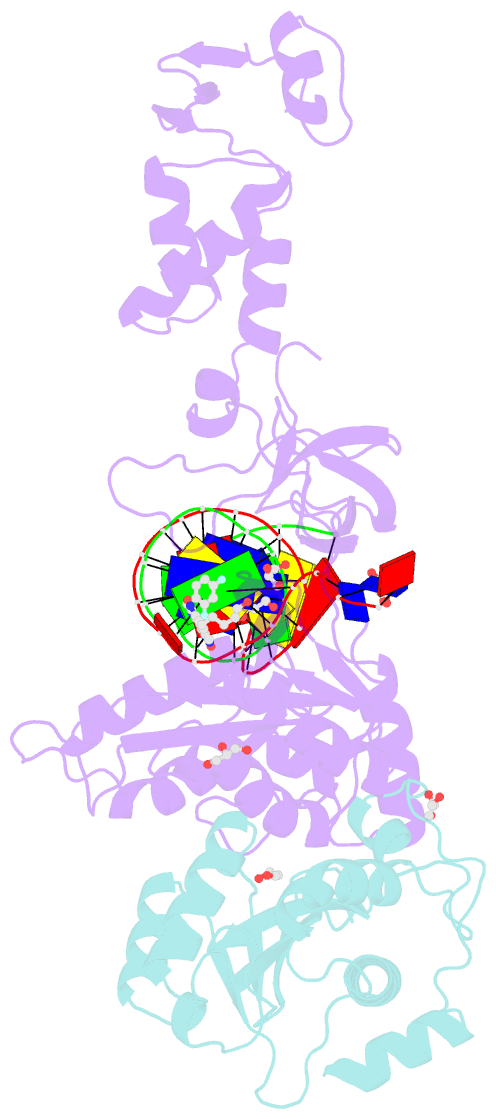
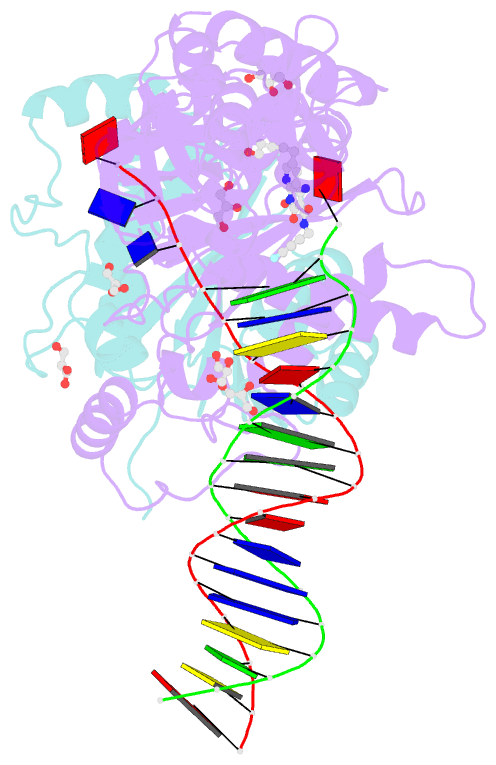
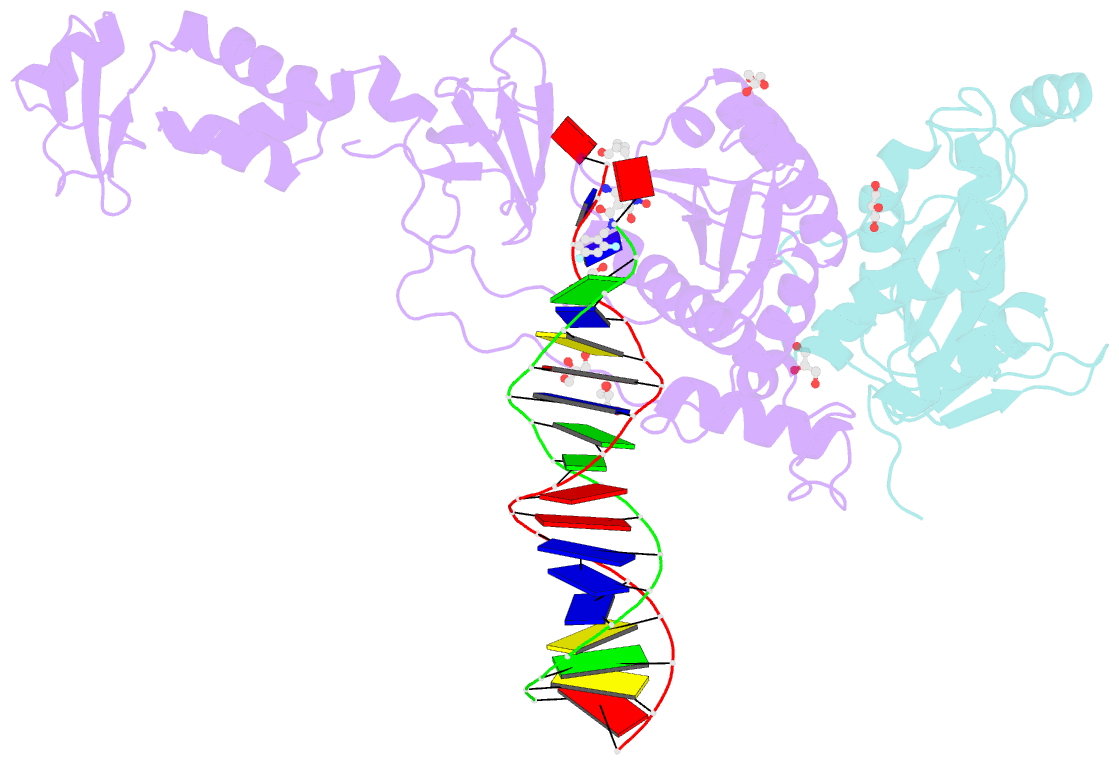
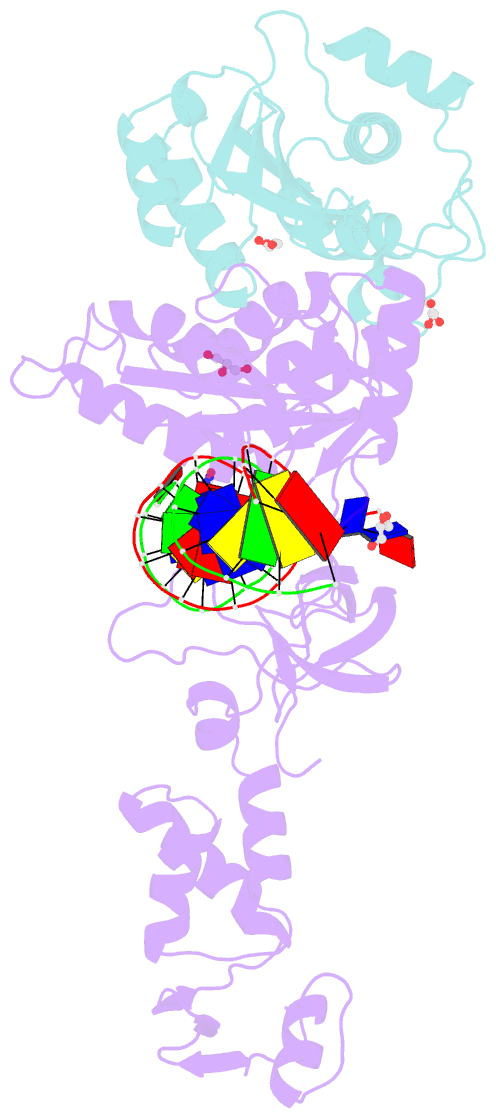
Summary information and primary citation
- PDB-id
- 5frn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- X-ray (2.85 Å)
- Summary
- Crystal structure of the prototype foamy virus (pfv) intasome in complex with magnesium and the insti xz419 (compound 4c)
- Reference
- Zhao XZ, Smith SJ, Maskell DP, Metifiot M, Pye VE, Fesen K, Marchand C, Pommier Y, Cherepanov P, Hughes SH, Burke TRJ (2016): "HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases." Acs Chem.Biol., 11, 1074. doi: 10.1021/ACSCHEMBIO.5B00948.
- Abstract
- HIV integrase (IN) strand transfer inhibitors (INSTIs) are among the newest anti-AIDS drugs; however, mutant forms of IN can confer resistance. We developed noncytotoxic naphthyridine-containing INSTIs that retain low nanomolar IC50 values against HIV-1 variants harboring all of the major INSTI-resistant mutations. We found by analyzing crystal structures of inhibitors bound to the IN from the prototype foamy virus (PFV) that the most successful inhibitors show striking mimicry of the bound viral DNA prior to 3'-processing and the bound host DNA prior to strand transfer. Using this concept of "bi-substrate mimicry," we developed a new broadly effective inhibitor that not only mimics aspects of both the bound target and viral DNA but also more completely fills the space they would normally occupy. Maximizing shape complementarity and recapitulating structural components encompassing both of the IN DNA substrates could serve as a guiding principle for the development of new INSTIs.