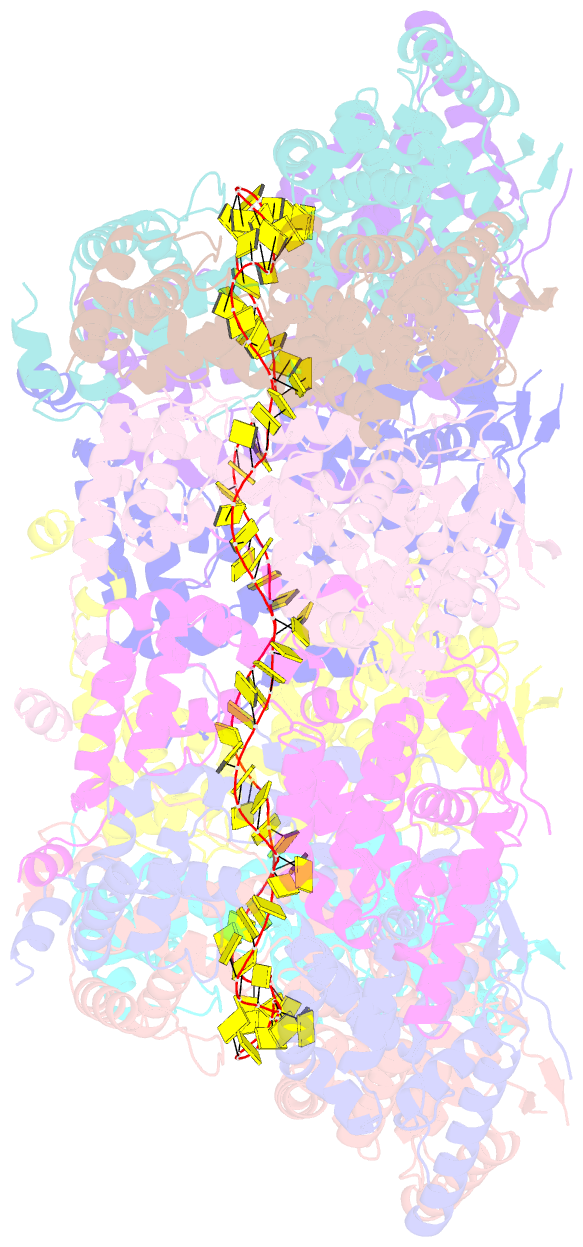
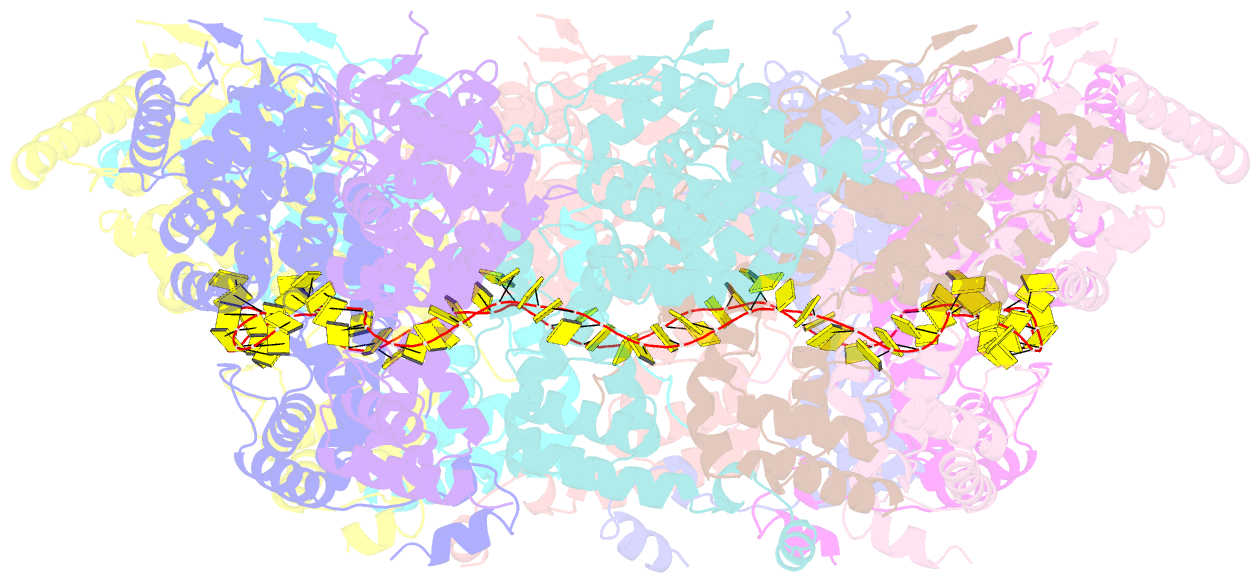
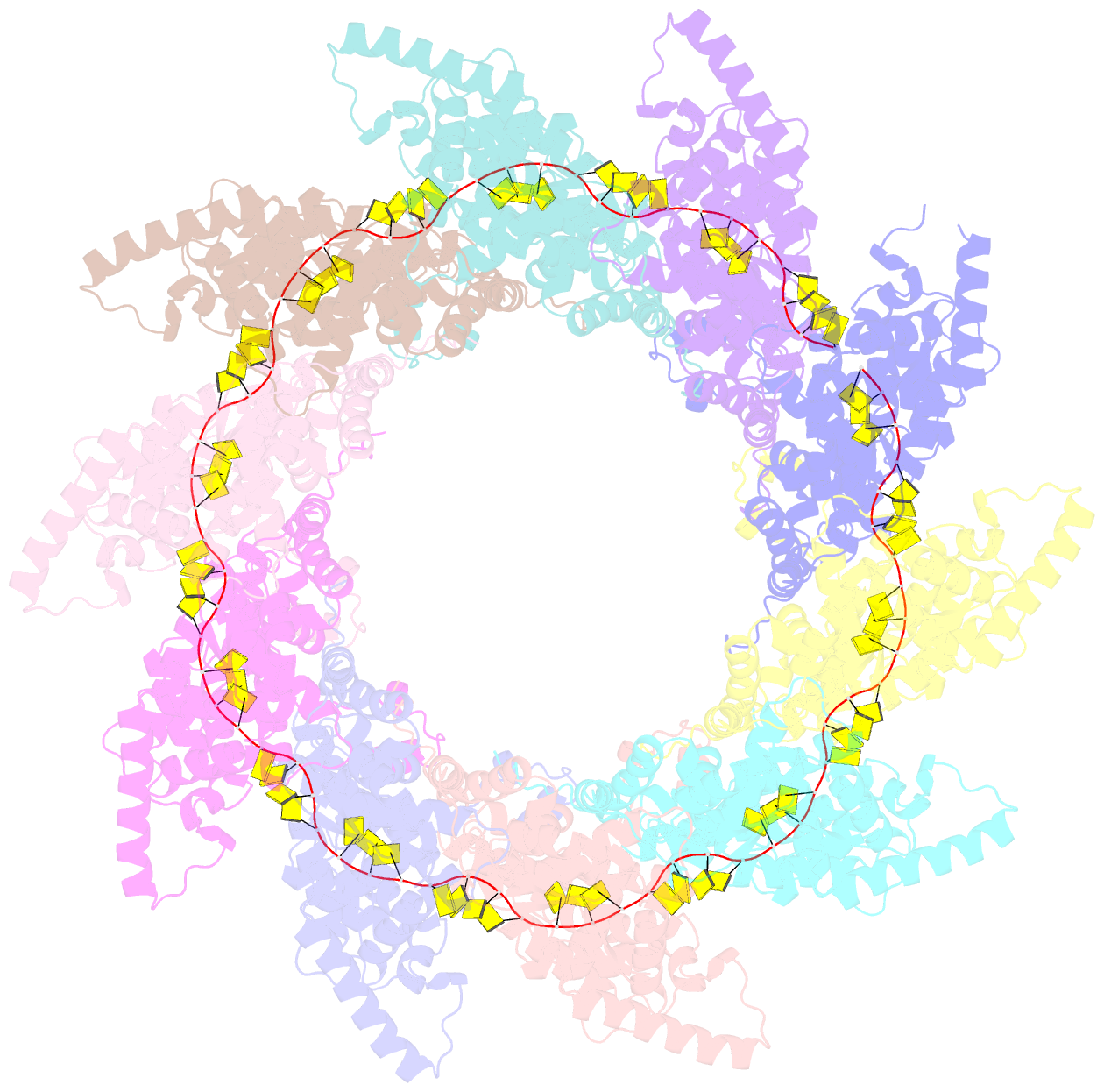
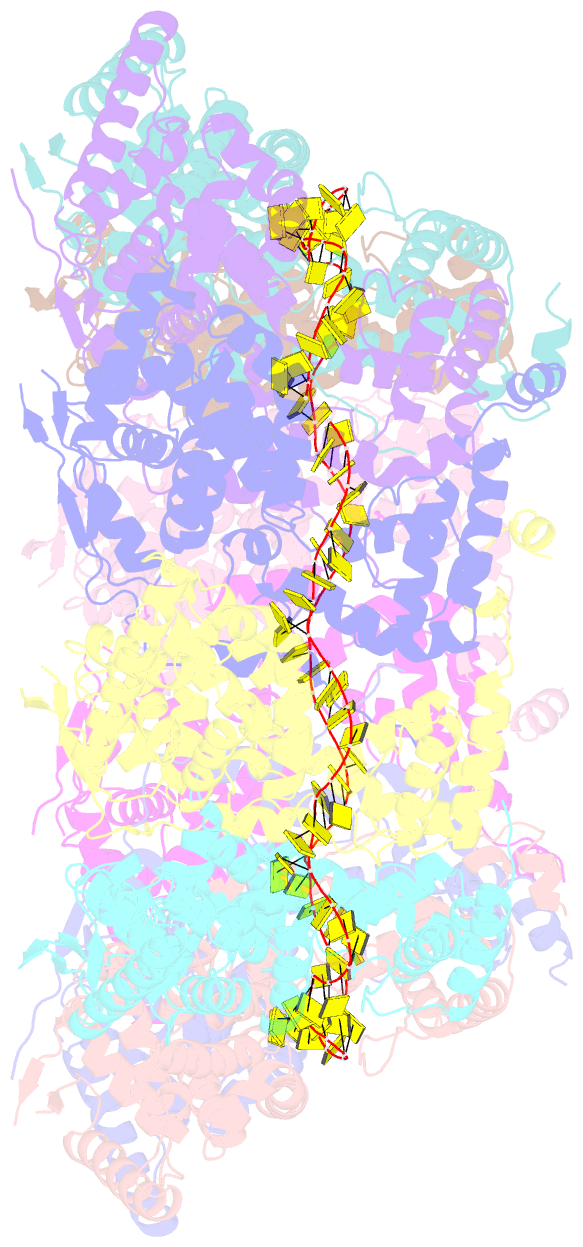
Summary information and primary citation
- PDB-id
- 5fvc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- X-ray (4.17 Å)
- Summary
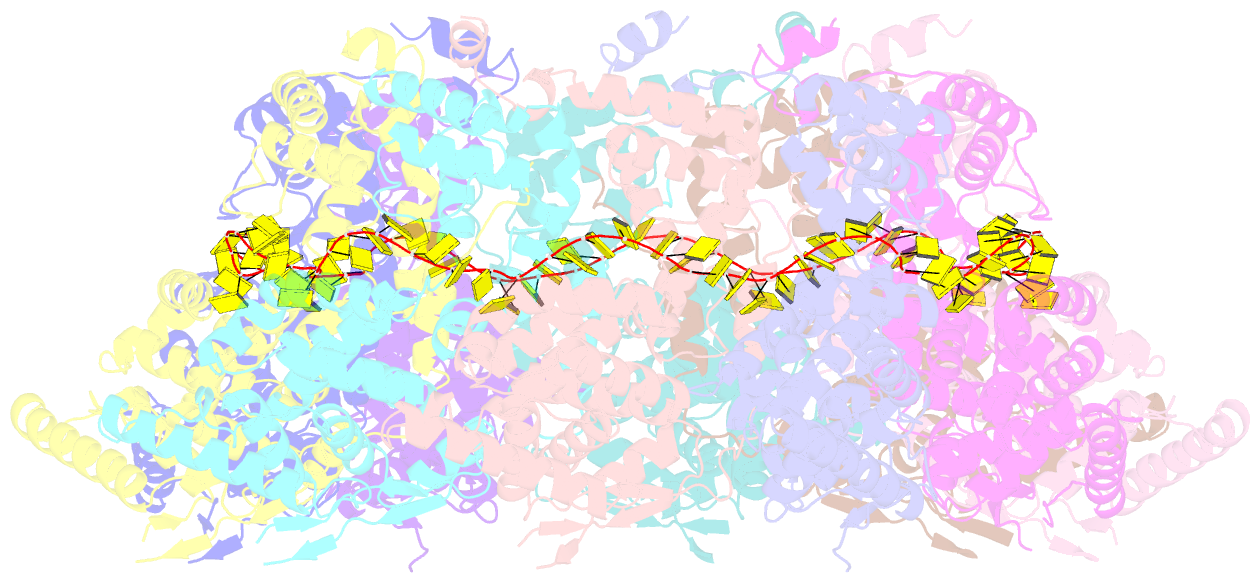
- Structure of RNA-bound decameric hmpv nucleoprotein
- Reference
- Renner M, Bertinelli M, Leyrat C, Paesen GC, Saraiva de Oliveira LF, Huiskonen JT, Grimes JM (2016): "Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein." Elife, 5, e12627. doi: 10.7554/eLife.12627.
- Abstract
- Non-segmented, (-)RNA viruses cause serious human diseases. Human metapneumovirus (HMPV), an emerging pathogen of this order of viruses (Mononegavirales) is one of the main causes of respiratory tract illness in children. To help elucidate the assembly mechanism of the nucleocapsid (the viral RNA genome packaged by the nucleoprotein N) we present crystallographic structures of HMPV N in its assembled RNA-bound state and in a monomeric state, bound to the polymerase cofactor P. Our structures reveal molecular details of how P inhibits the self-assembly of N and how N transitions between the RNA-free and RNA-bound conformational state. Notably, we observe a role for the C-terminal extension of N in directly preventing premature uptake of RNA by folding into the RNA-binding cleft. Our structures suggest a common mechanism of how the growth of the nucleocapsid is orchestrated, and highlight an interaction site representing an important target for antivirals.