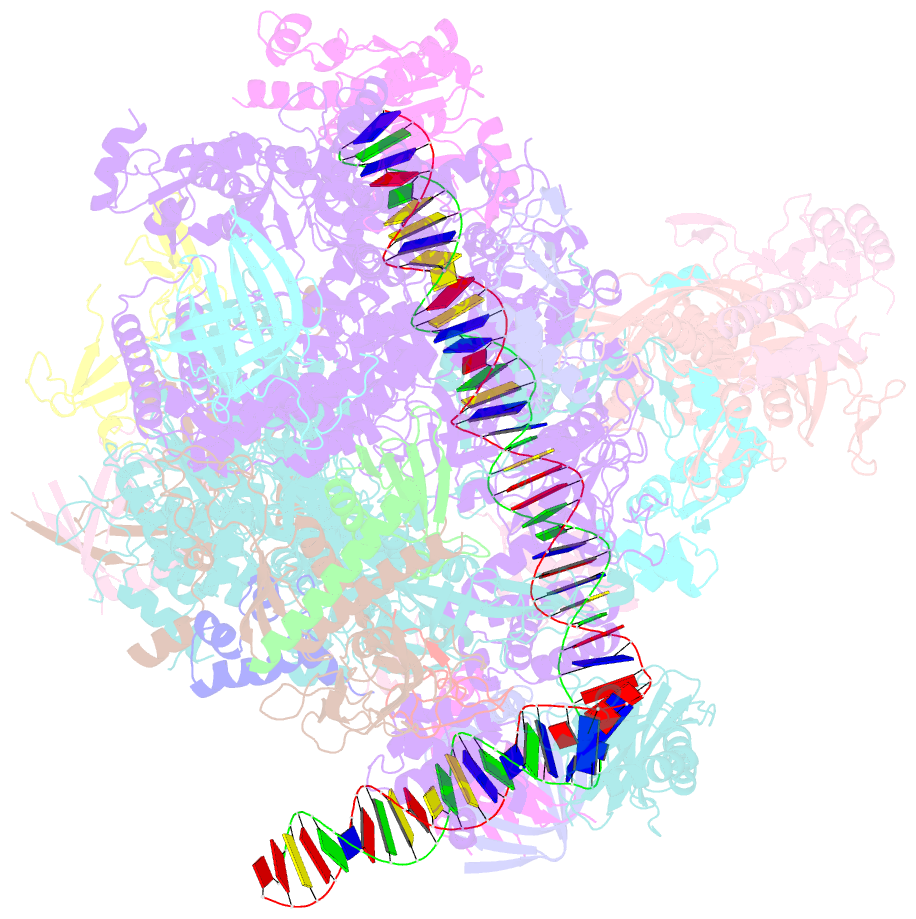
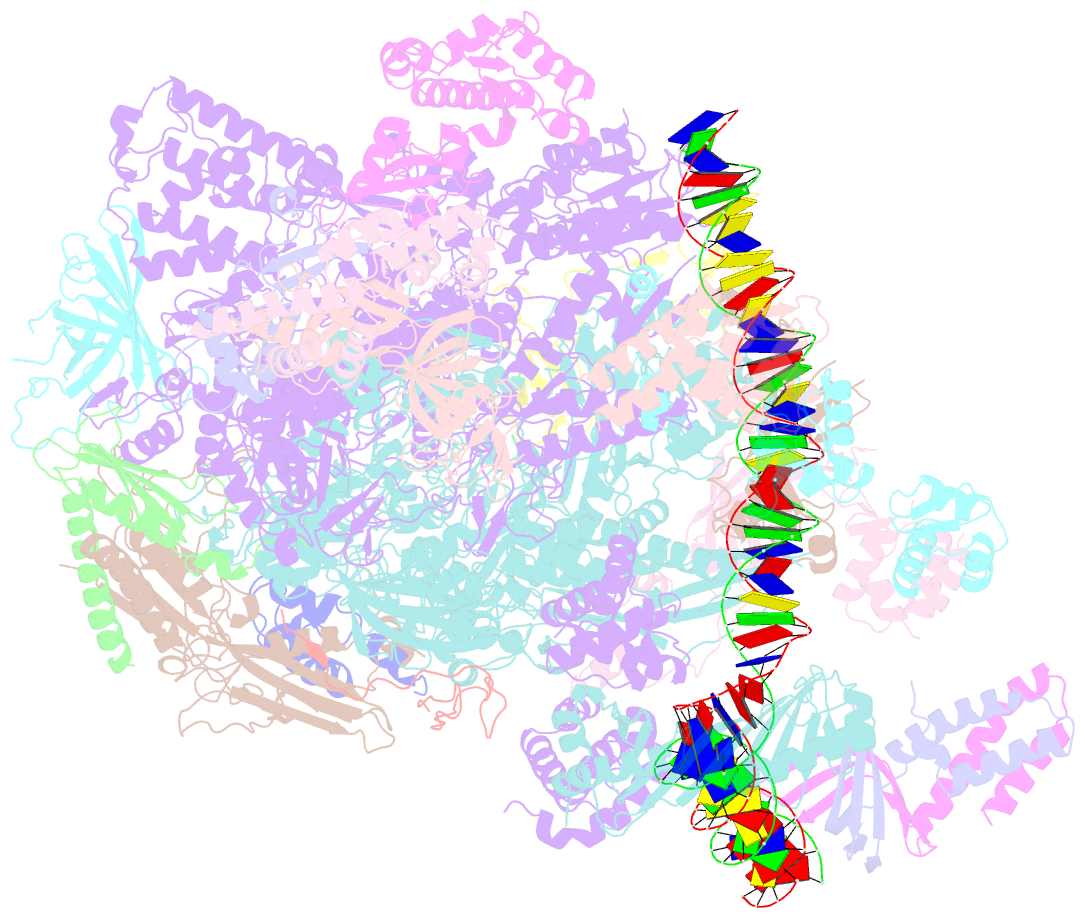
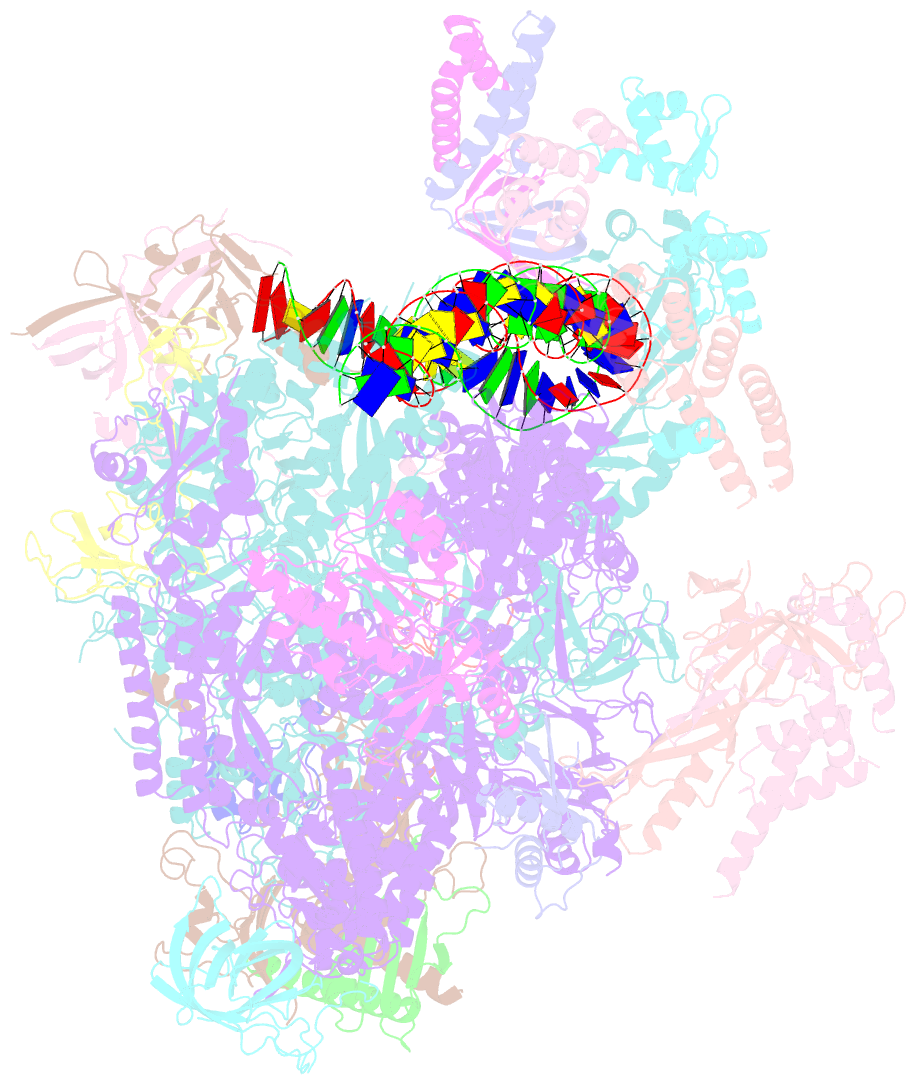
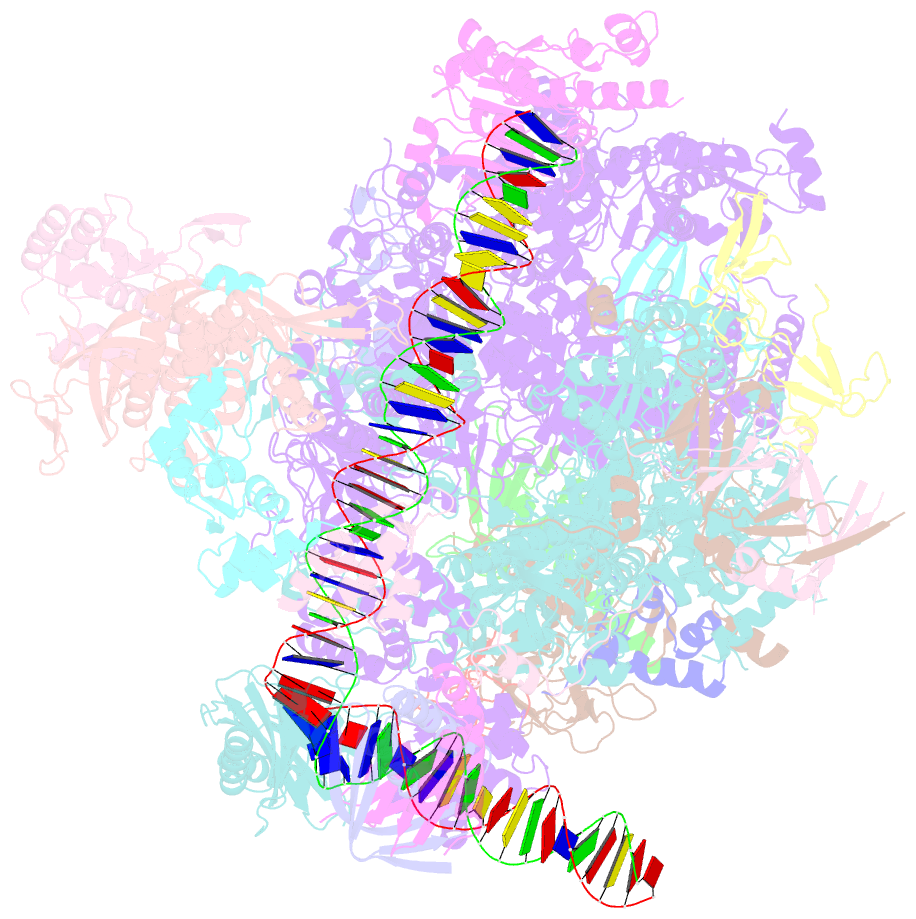
Summary information and primary citation
- PDB-id
- 5fz5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (8.8 Å)
- Summary
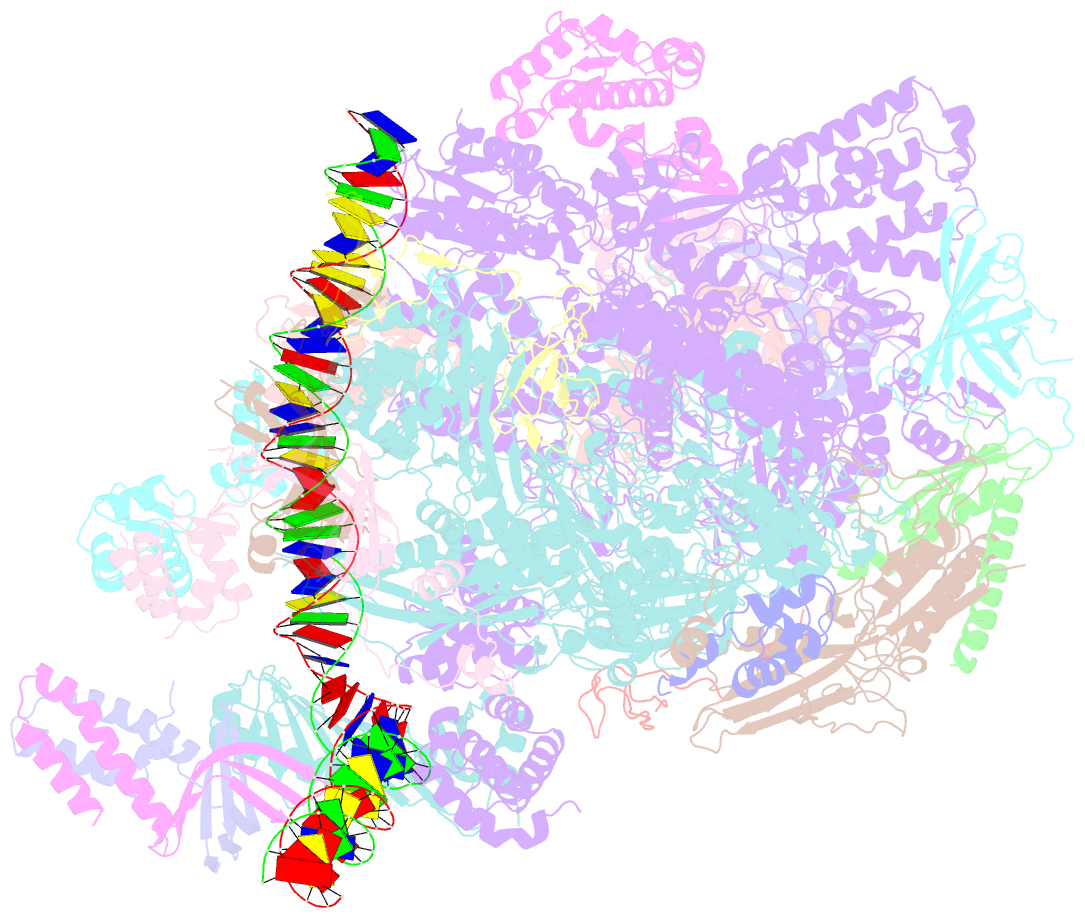
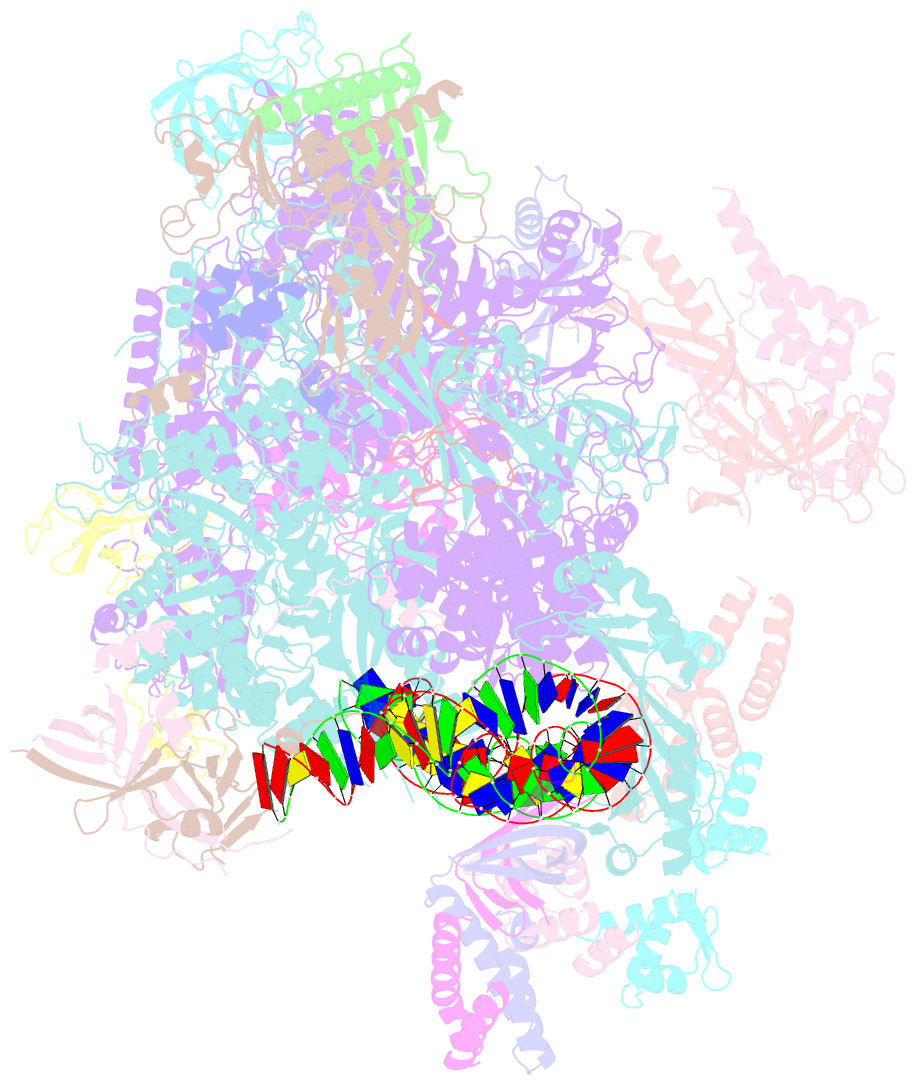
- Transcription initiation complex structures elucidate DNA opening (cc)
- Reference
- Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P (2016): "Transcription Initiation Complex Structures Elucidate DNA Opening." Nature, 533, 353. doi: 10.1038/NATURE17990.
- Abstract
- Transcription of eukaryotic protein-coding genes begins with assembly of the RNA polymerase (Pol) II initiation complex and promoter DNA opening. Here we report cryo-electron microscopy (cryo-EM) structures of yeast initiation complexes containing closed and open DNA at resolutions of 8.8 Å and 3.6 Å, respectively. DNA is positioned and retained over the Pol II cleft by a network of interactions between the TATA-box-binding protein TBP and transcription factors TFIIA, TFIIB, TFIIE, and TFIIF. DNA opening occurs around the tip of the Pol II clamp and the TFIIE 'extended winged helix' domain, and can occur in the absence of TFIIH. Loading of the DNA template strand into the active centre may be facilitated by movements of obstructing protein elements triggered by allosteric binding of the TFIIE 'E-ribbon' domain. The results suggest a unified model for transcription initiation with a key event, the trapping of open promoter DNA by extended protein-protein and protein-DNA contacts.