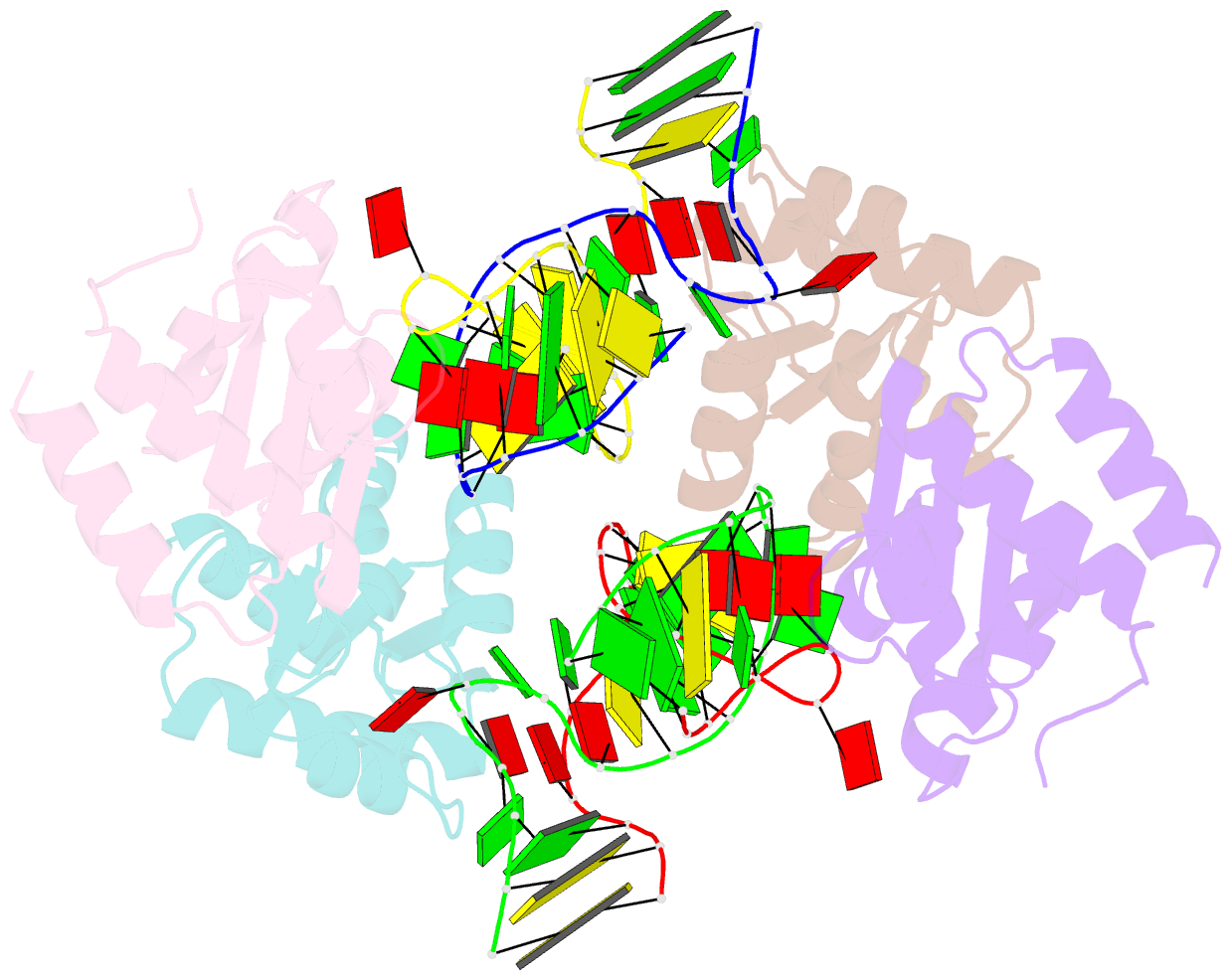
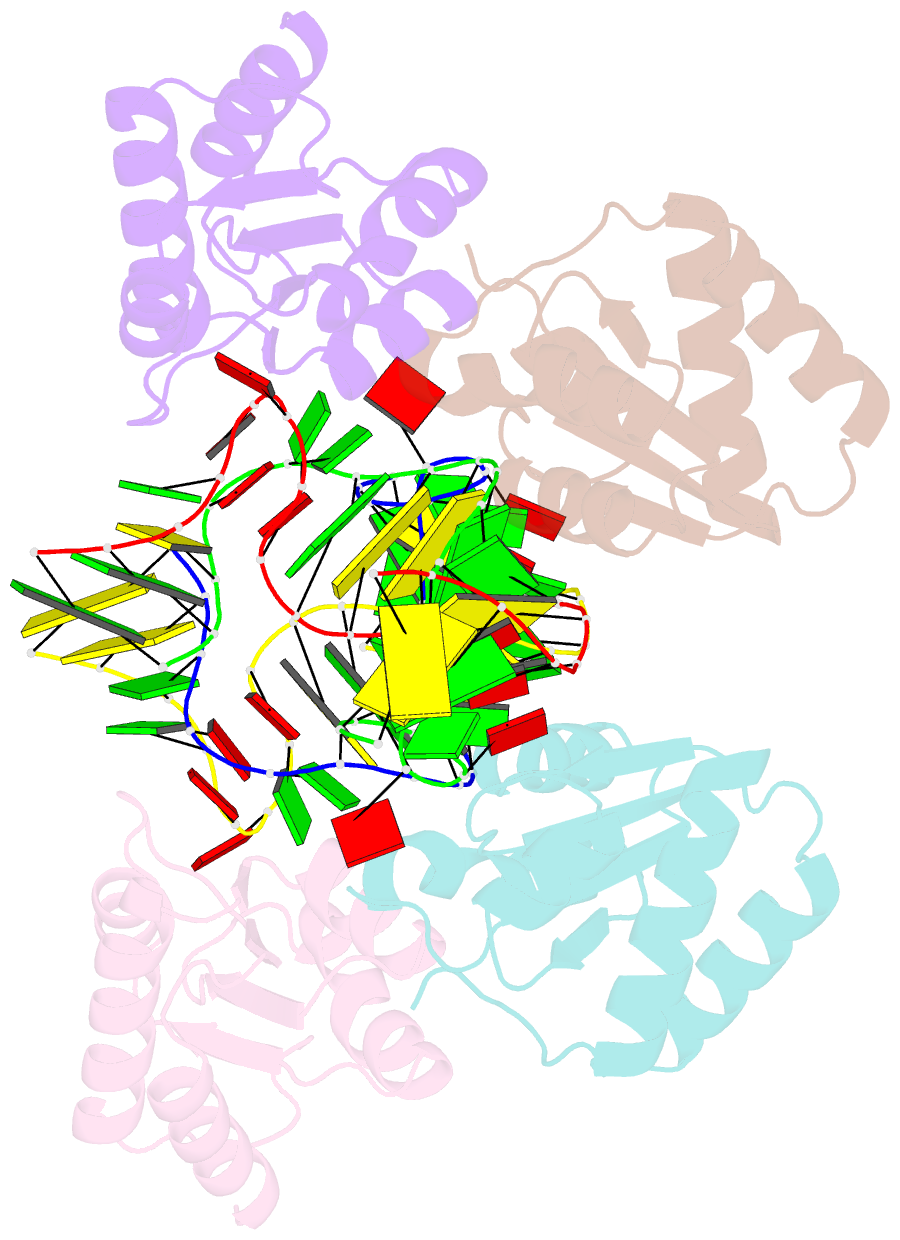
Summary information and primary citation
- PDB-id
- 5g4v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding
- Method
- X-ray (2.87 Å)
- Summary
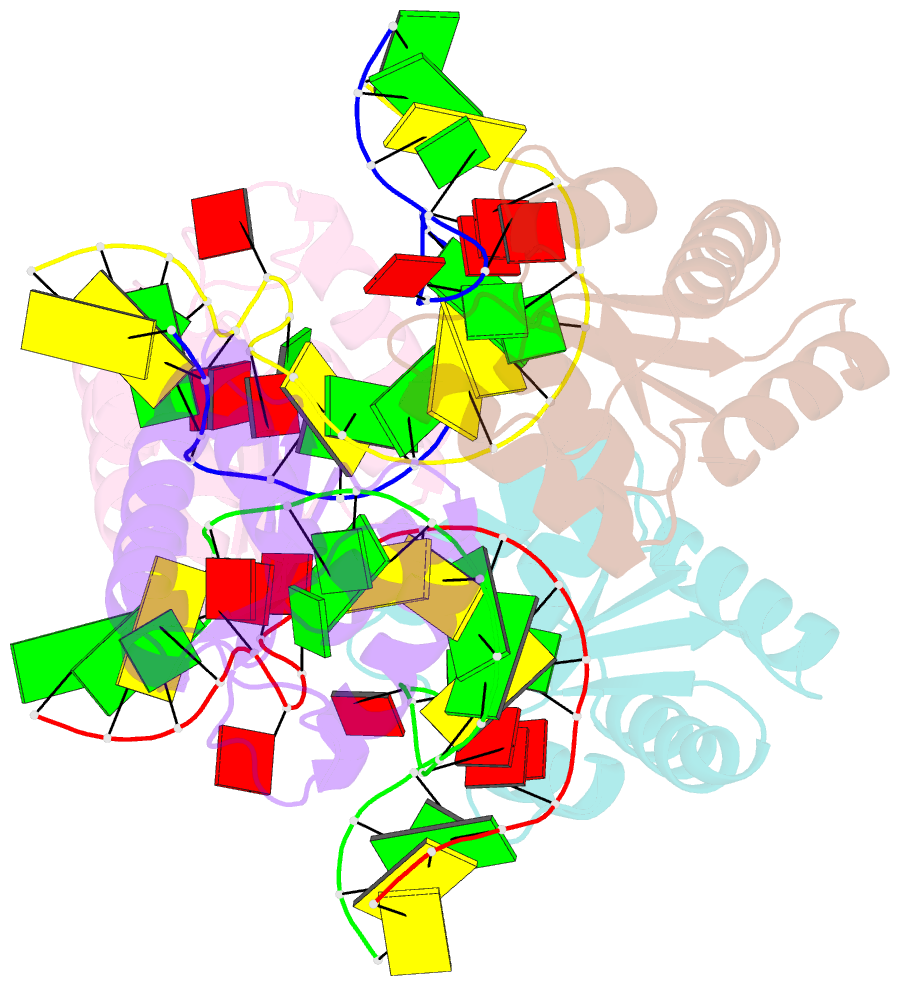
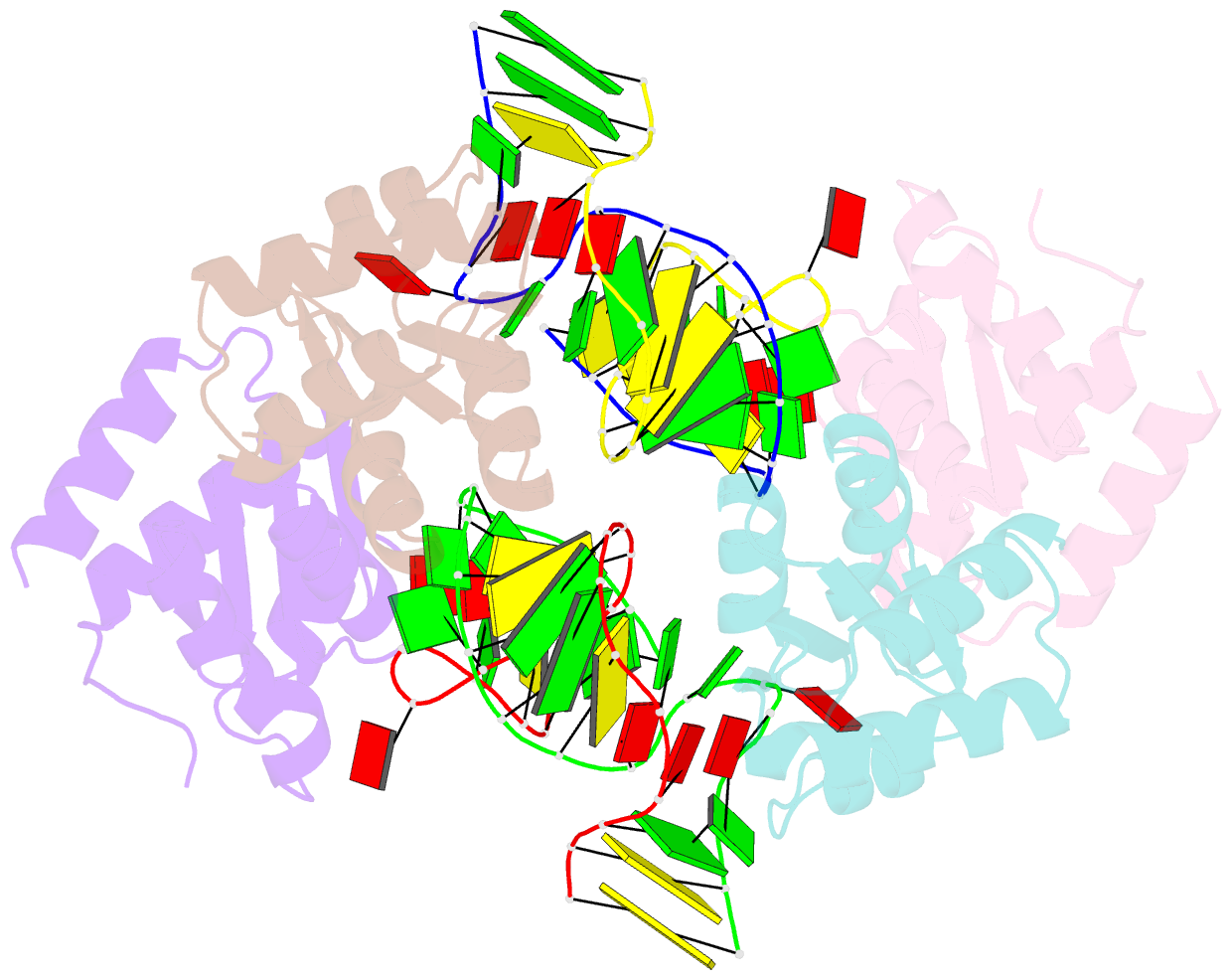
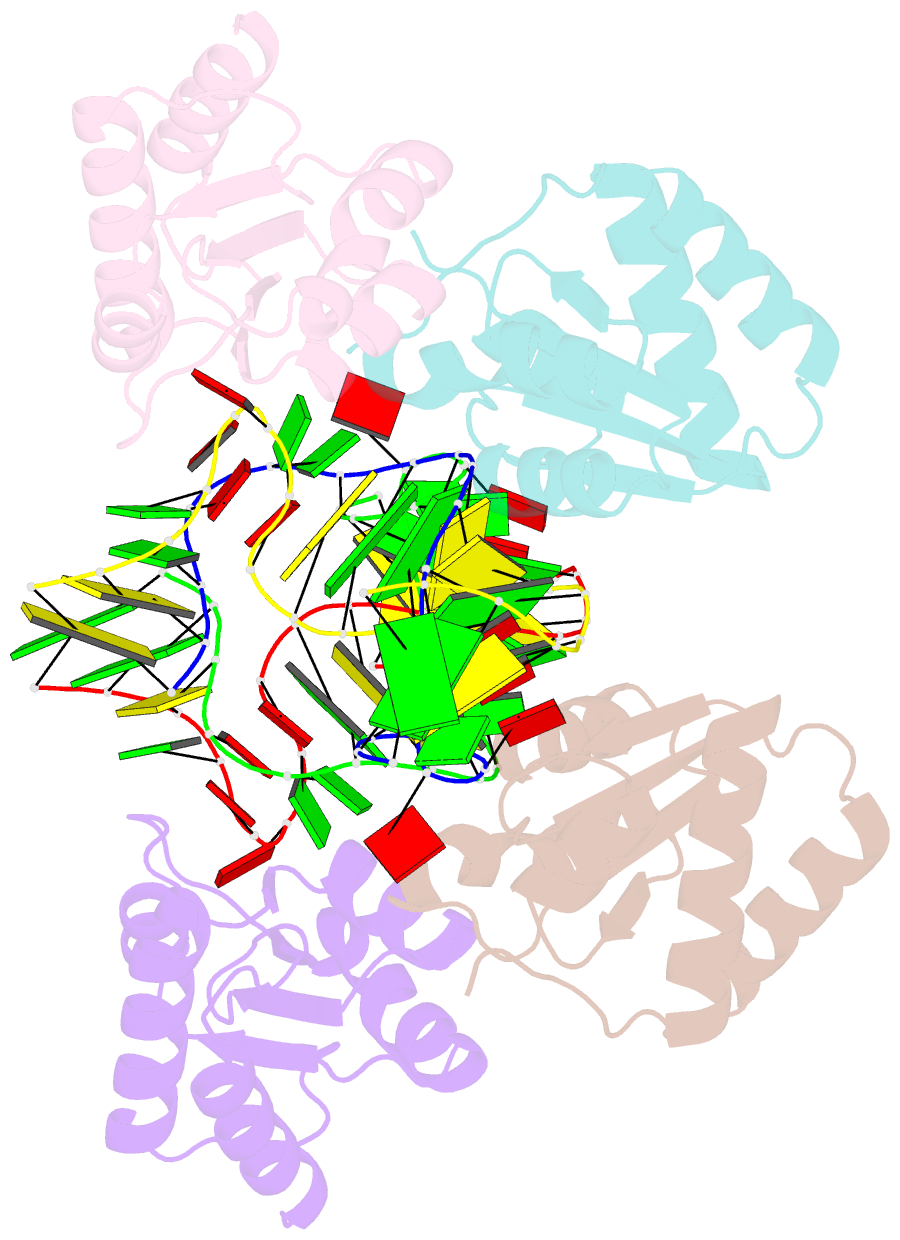
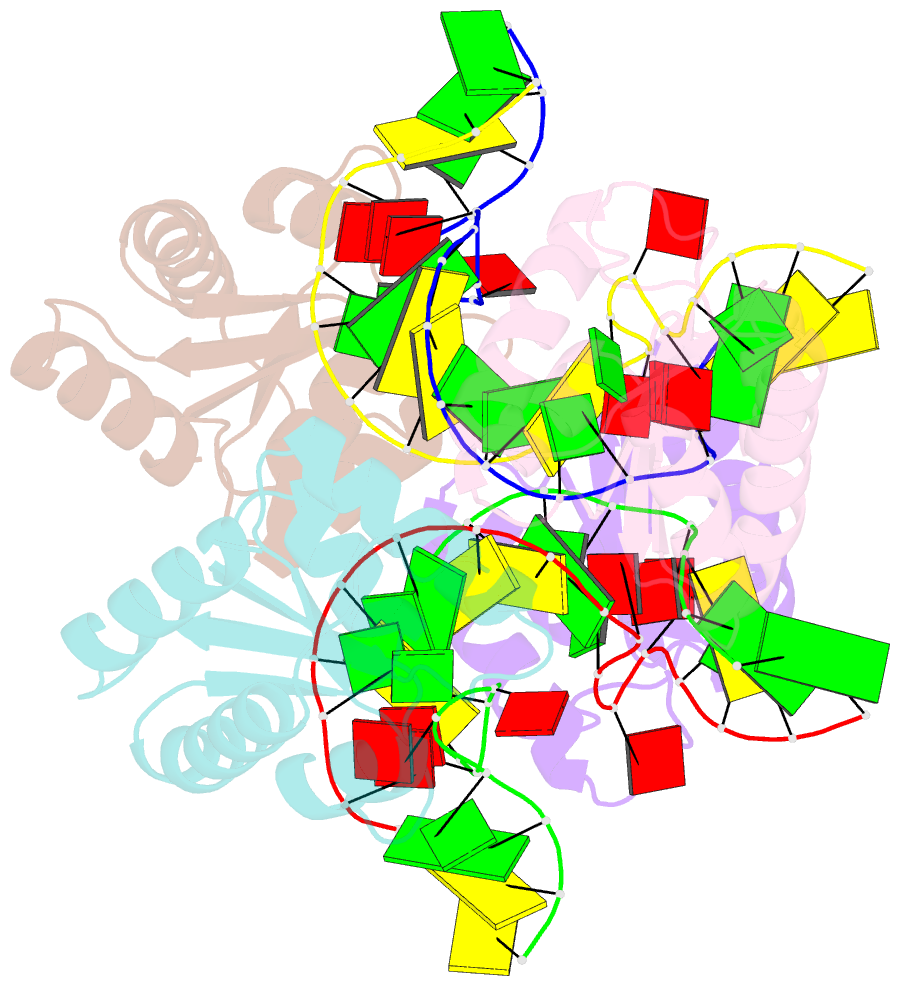
- Association of four two-k-turn units based on kt-7 3bg,3nc, forming a square-shaped structure
- Reference
- Huang L, Lilley DMJ (2016): "A Quasi-Cyclic RNA Nano-Scale Molecular Object Constructed Using Kink Turns." Nanoscale, 8, 15189. doi: 10.1039/C6NR05186C.
- Abstract
- k-Turns are widespread RNA architectural elements that mediate tertiary interactions. We describe a double-kink-turn motif comprising two inverted k-turns that forms a tight horse-shoe structure that can assemble into a variety of shapes by coaxial association of helical ends. Using X-ray crystallography we show that these assemble with two (dumbell), three (triangle) and four units (square), with or without bound protein, within the crystal lattice. In addition, exchange of a single basepair can almost double the pore radius or shape of a molecular assembly. On the basis of this analysis we synthesized a 114 nt self-complementary RNA containing six k-turns. The crystal structure of this species shows that it forms a quasi-cyclic triangular object. These are randomly disposed about the three-fold axis in the crystal lattice, generating a circular RNA of quasi D3 symmetry with a shape reminiscent of that of a cyclohexane molecule in its chair conformation. This work demonstrates that the k-turn is a powerful building block in the construction of nano-scale molecular objects, and illustrates why k-turns are widely used in natural RNA molecules to organize long-range architecture and mediate tertiary contacts.