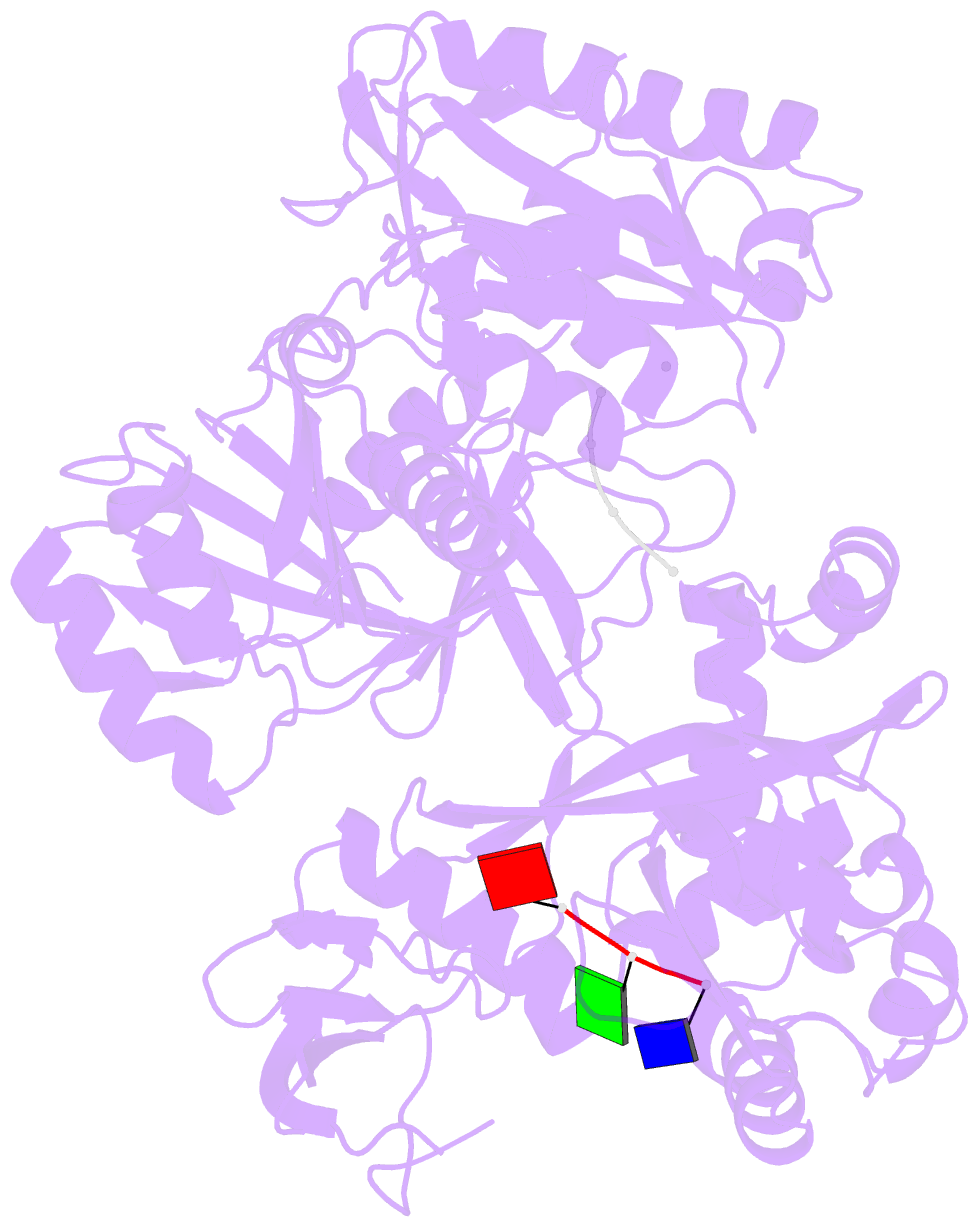
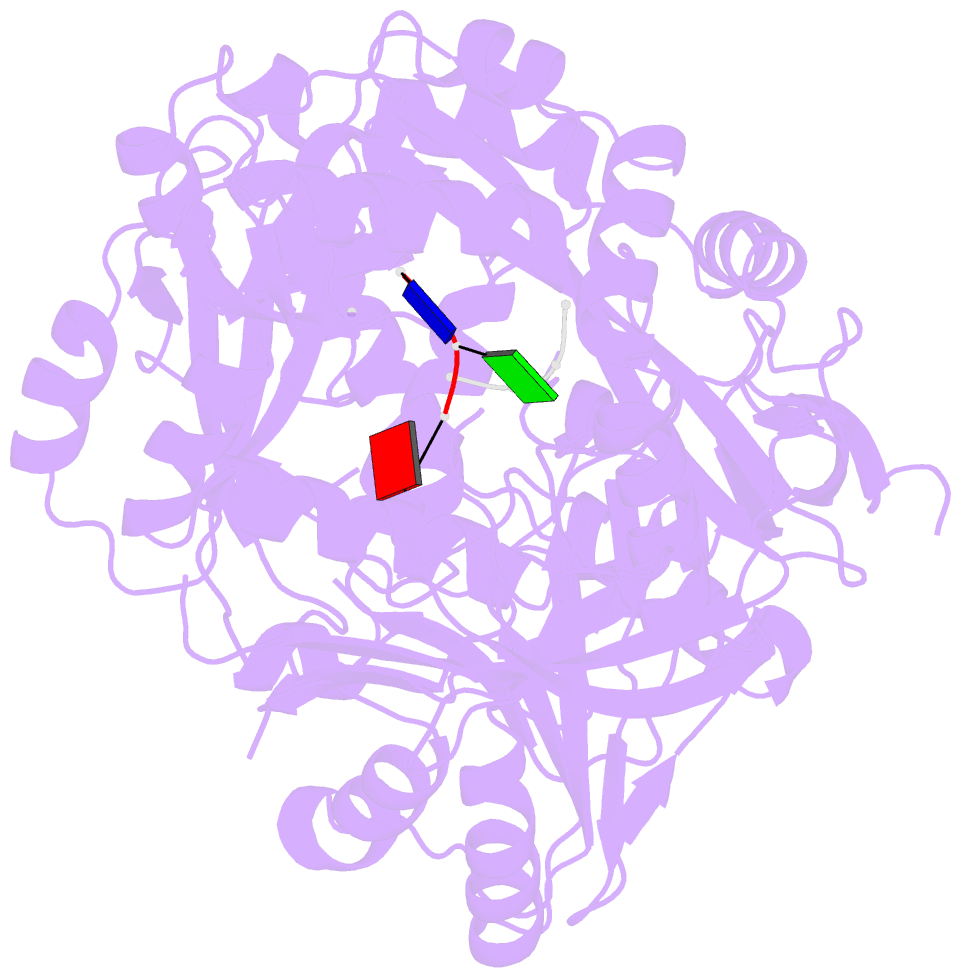
Summary information and primary citation
- PDB-id
- 5g5t; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.85 Å)
- Summary
- Structure of the argonaute protein from methanocaldcoccus janaschii in complex with guide DNA
- Reference
- Willkomm S, Oellig CA, Zander A, Restle T, Keegan R, Grohmann D, Schneider S (2017): "Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein." Nat Microbiol, 2, 17035. doi: 10.1038/nmicrobiol.2017.35.
- Abstract
- Argonaute (Ago) proteins in eukaryotes are known as key players in post-transcriptional gene silencing1, while recent studies on prokaryotic Agos hint at their role in the protection against invading DNA2,3. Here, we present crystal structures of the apo enzyme and a binary Ago-guide complex of the archaeal Methanocaldococcus jannaschii (Mj) Ago. Binding of a guide DNA leads to large structural rearrangements. This includes the structural transformation of a hinge region containing a switch helix, which has been shown for human Ago2 to be critical for the dynamic target search process4-6. To identify key residues crucial for MjAgo function, we analysed the effect of several MjAgo mutants. We observe that the nature of the 3' and 5' nucleotides in particular, as well as the switch helix, appear to impact MjAgo cleavage activity. In summary, we provide insights into the molecular mechanisms that drive DNA-guided DNA silencing by an archaeal Ago.