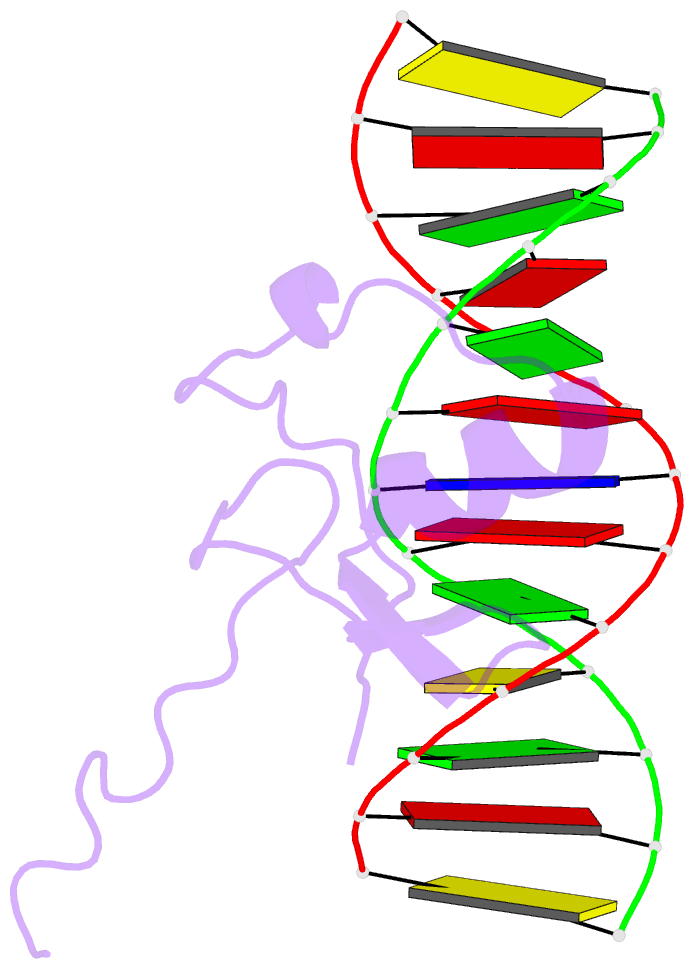
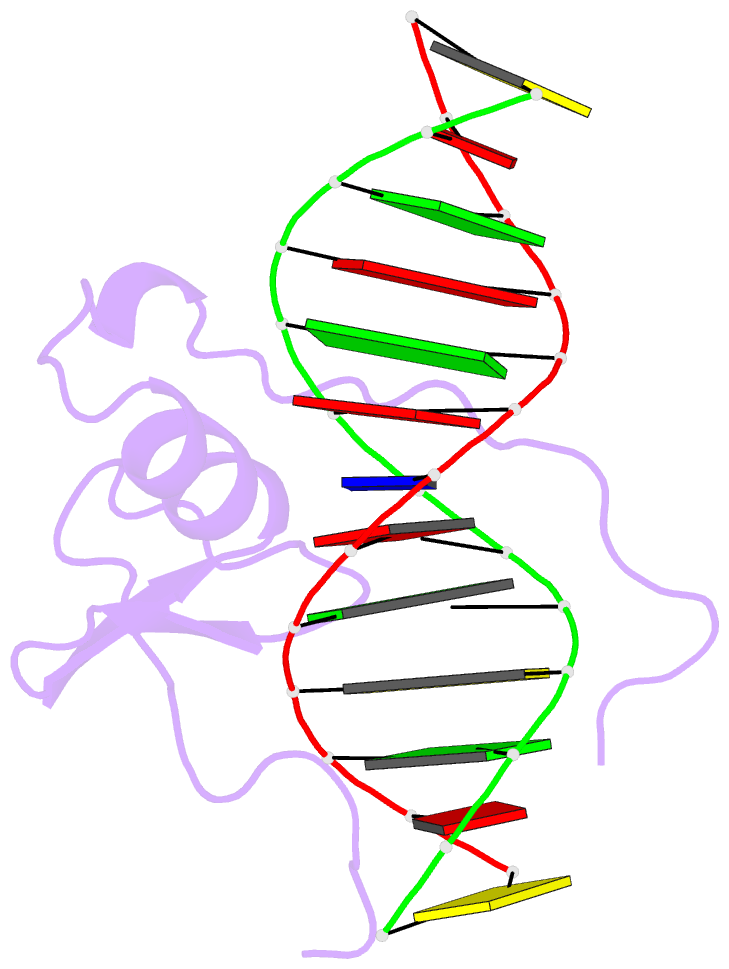
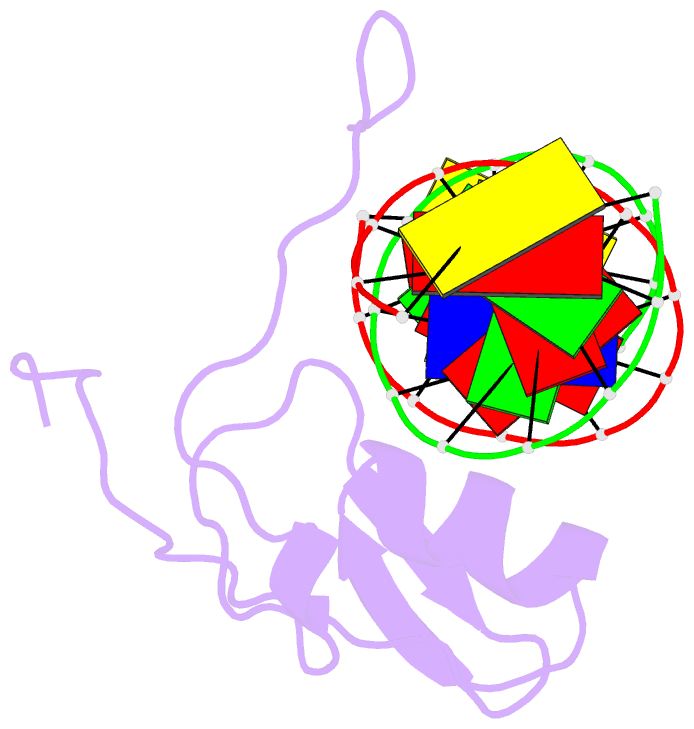
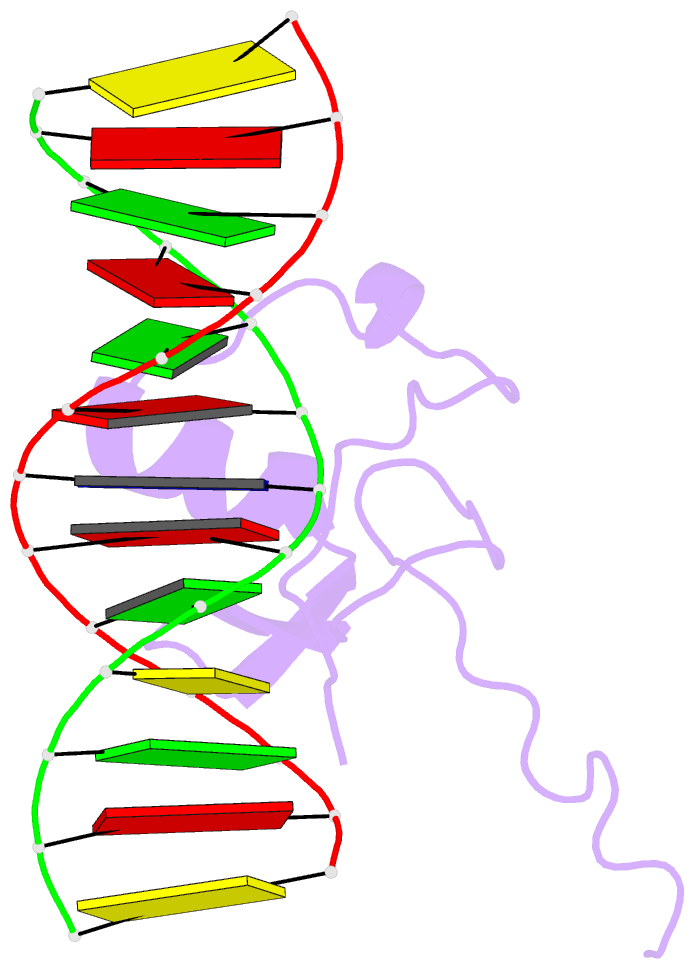
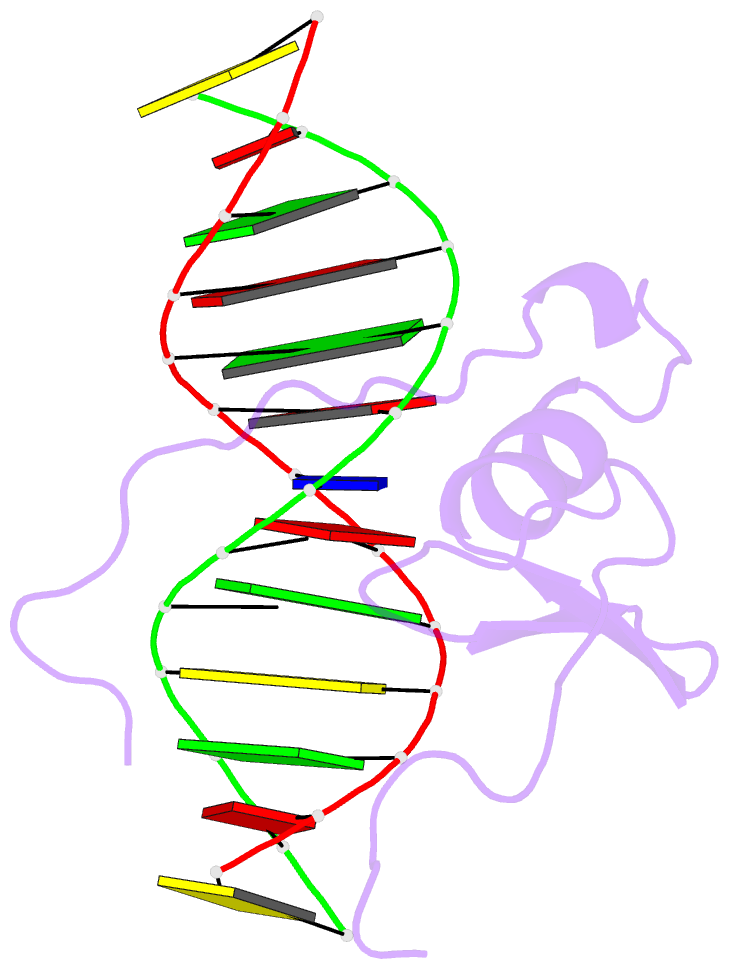
Summary information and primary citation
- PDB-id
- 5gat; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- NMR
- Summary
- Solution NMR structure of the wild type DNA binding domain of area complexed to a 13bp DNA containing a cgata site, 35 structures
- Reference
- Starich MR, Wikstrom M, Arst Jr HN, Clore GM, Gronenborn AM (1998): "The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors." J.Mol.Biol., 277, 605-620. doi: 10.1006/jmbi.1998.1625.
- Abstract
- The solution structure of a complex between the DNA binding domain of a fungal GATA factor and a 13 base-pair oligonucleotide containing its physiologically relevant CGATAG target sequence has been determined by multidimensional nuclear magnetic resonance spectroscopy. The AREA DNA binding domain, from Aspergillus nidulans, possesses a single Cys2-Cys2 zinc finger module and a basic C-terminal tail, which recognize the CGATAG element via an extensive network of hydrophobic interactions with the bases in the major groove and numerous non-specific contacts along the sugar-phosphate backbone. The zinc finger core of the AREA DNA binding domain has the same global fold as that of the C-terminal DNA binding domain of chicken GATA-1. In contrast to the complex with the DNA binding domain of GATA-1 in which the basic C-terminal tail wraps around the DNA and lies in the minor groove, the structure of complex with the AREA DNA binding domain reveals that the C-terminal tail of the fungal domain runs parallel with the sugar phosphate backbone along the edge of the minor groove. This difference is principally attributed to amino acid substitutions at two positions of the AREA DNA binding domain (Val55, Asn62) relative to that of GATA-1 (Gly55, Lys62). The impact of the different C-terminal tail binding modes on the affinity and specificity of GATA factors is discussed.