Summary information and primary citation
- PDB-id
- 5gpc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription, DNA binding protein-DNA
- Method
- X-ray (2.8 Å)
- Summary
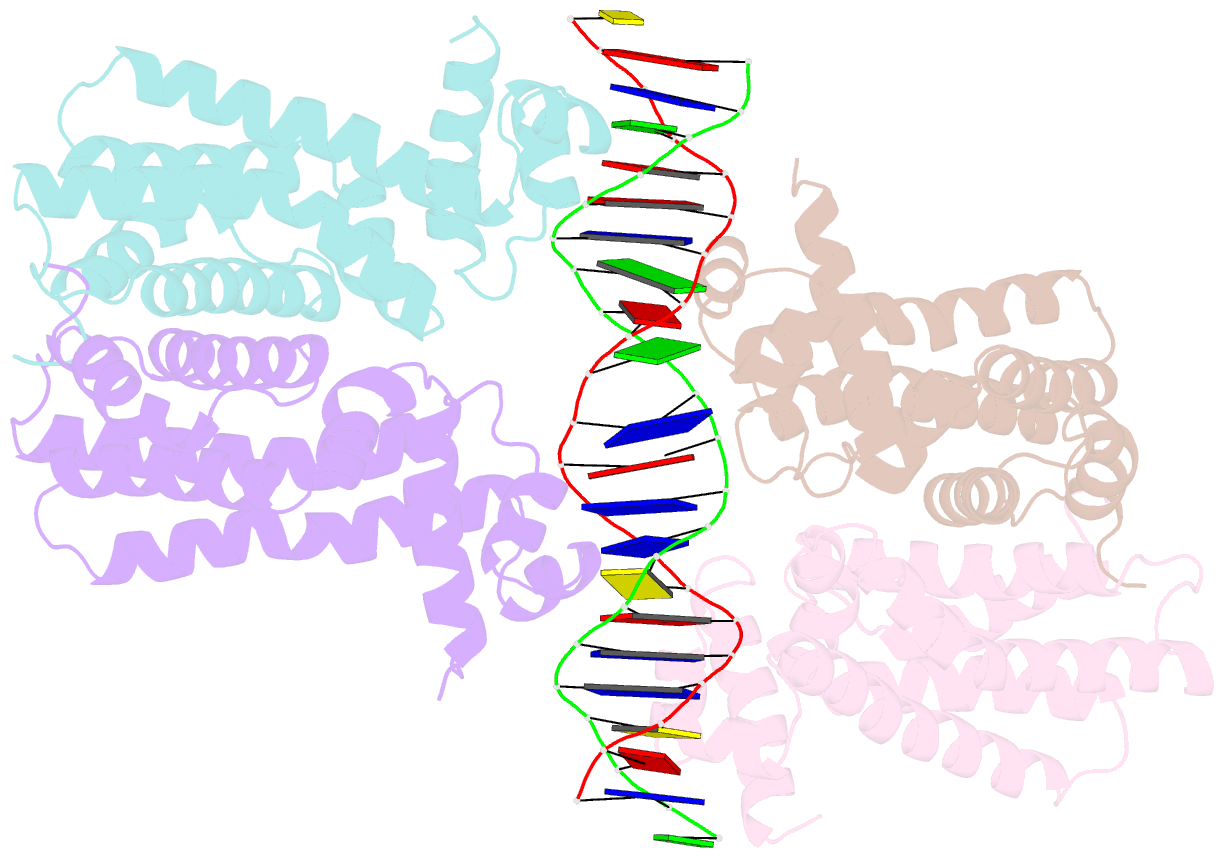
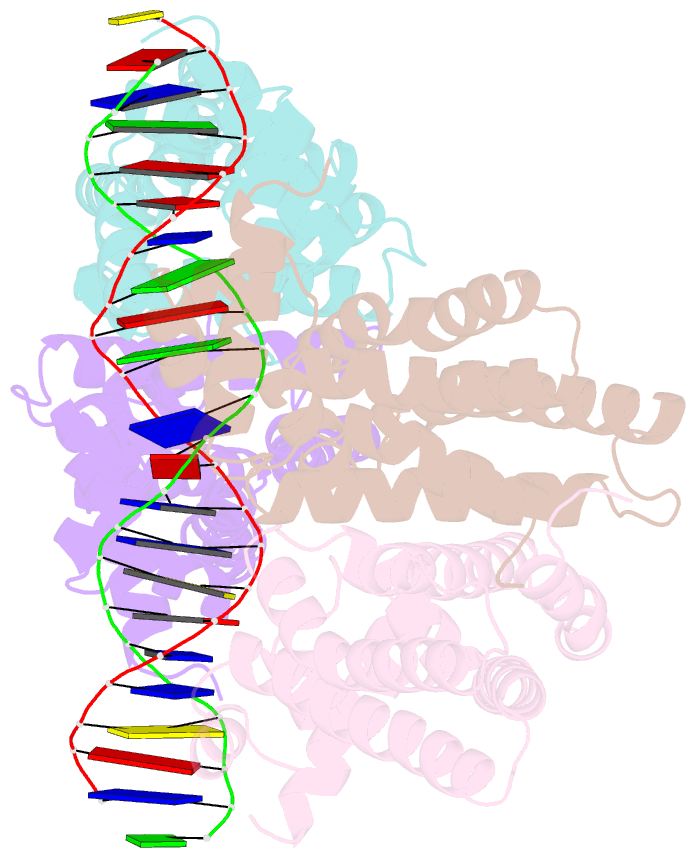
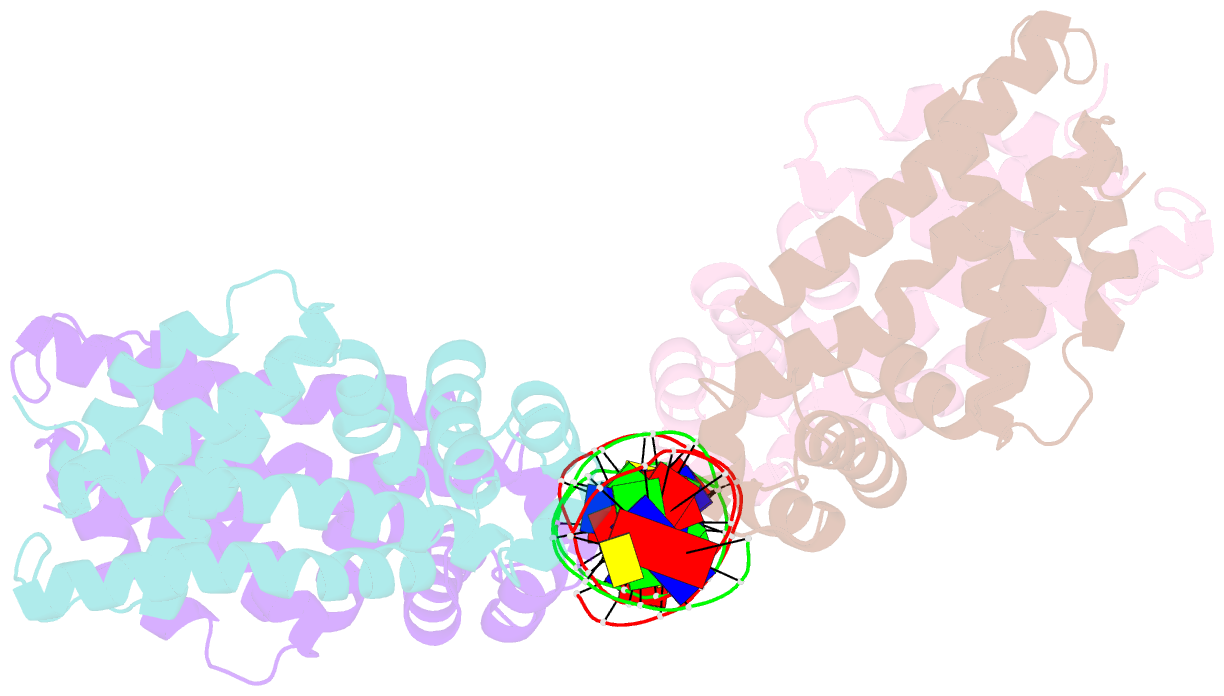
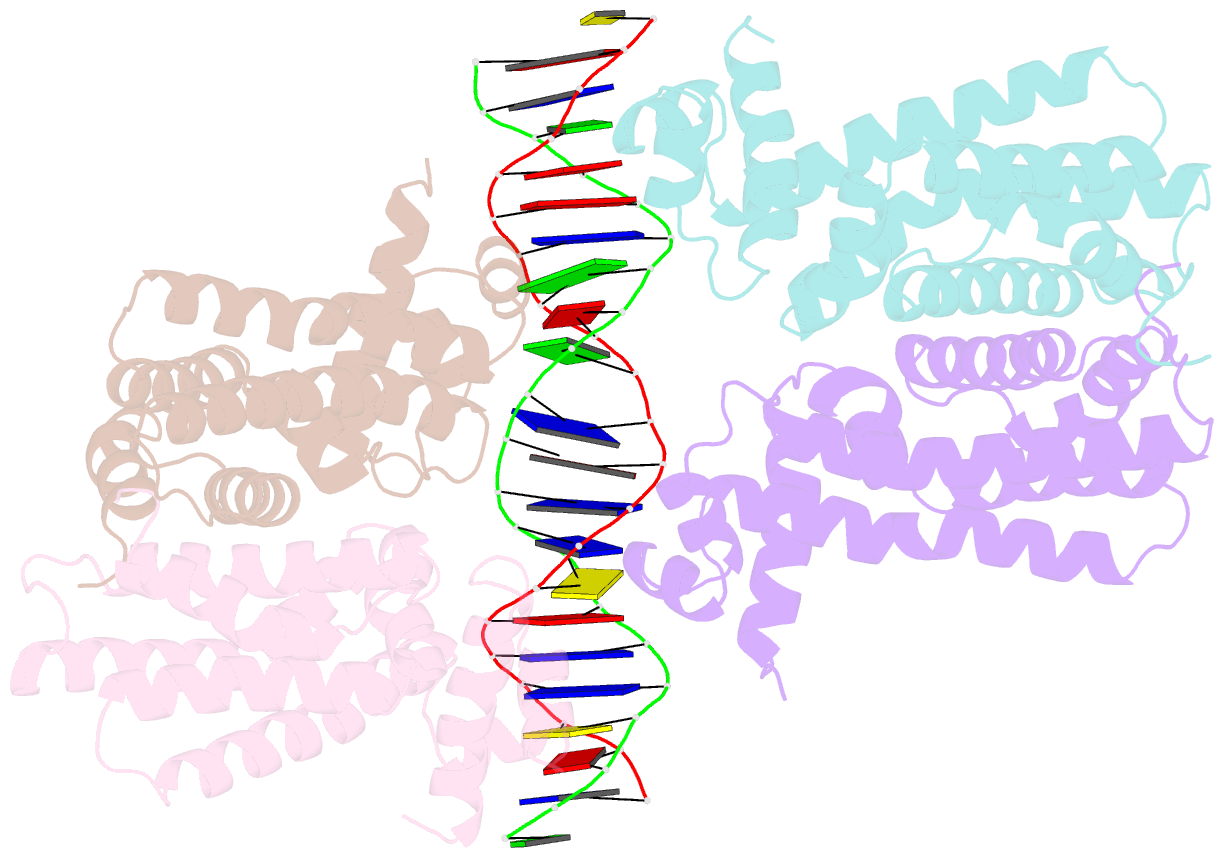
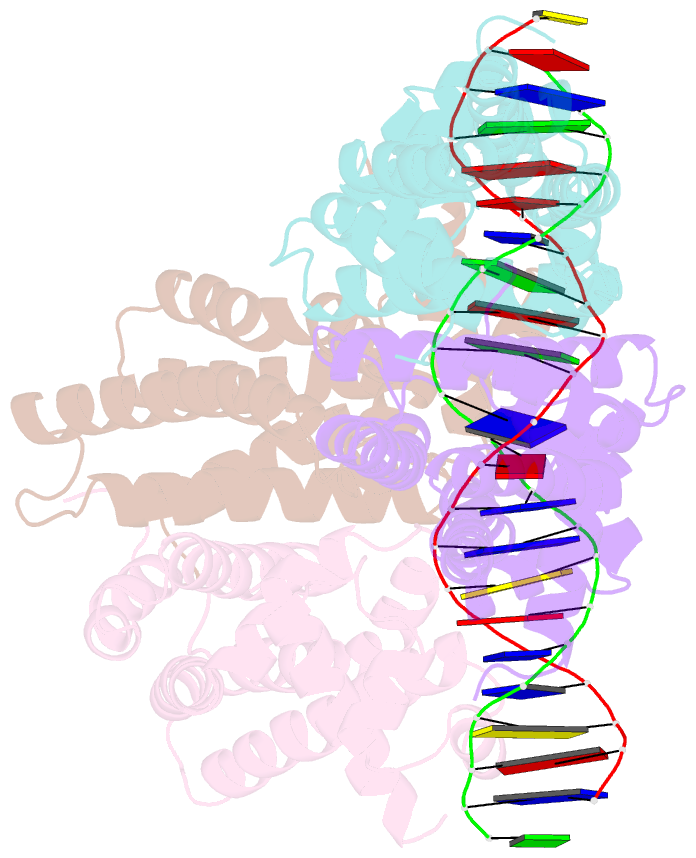
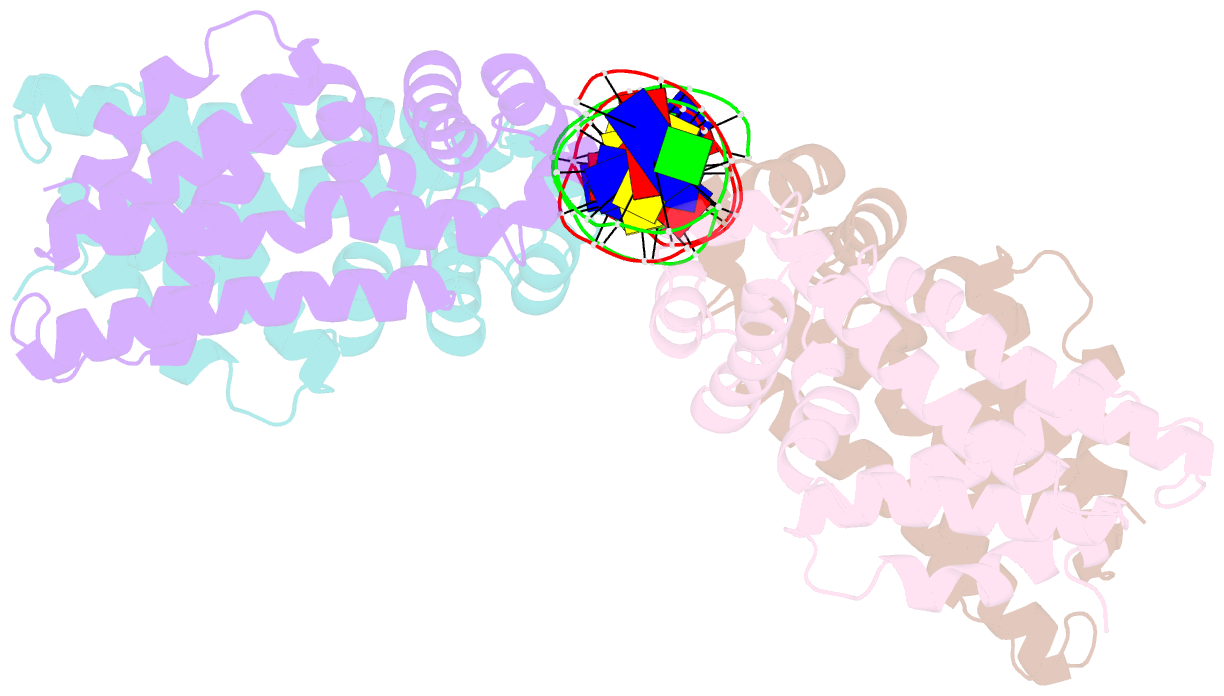
- Structural analysis of fatty acid degradation regulator fadr from bacillus halodurans
- Reference
- Yeo HK, Park YW, Lee JY (2017): "Structural basis of operator sites recognition and effector binding in the TetR family transcription regulator FadR." Nucleic Acids Res., 45, 4244-4254. doi: 10.1093/nar/gkx009.
- Abstract
- FadR is a fatty acyl-CoA dependent transcription factor that regulates genes encoding proteins involved in fatty-acid degradation and synthesis pathways. In this study, the crystal structures of Bacillus halodurans FadR, which belong to the TetR family, have been determined in three different forms: ligand-bound, ligand-free and DNA-bound at resolutions of 1.75, 2.05 and 2.80 Å, respectively. Structural and functional data showed that B. halodurans FadR was bound to its operator site without fatty acyl-CoAs. Structural comparisons among the three different forms of B. halodurans FadR revealed that the movement of DNA binding domains toward the operator DNA was blocked upon binding of ligand molecules. These findings suggest that the TetR family FadR negatively regulates the genes involved in fatty acid metabolism by binding cooperatively to the operator DNA as a dimer of dimers.