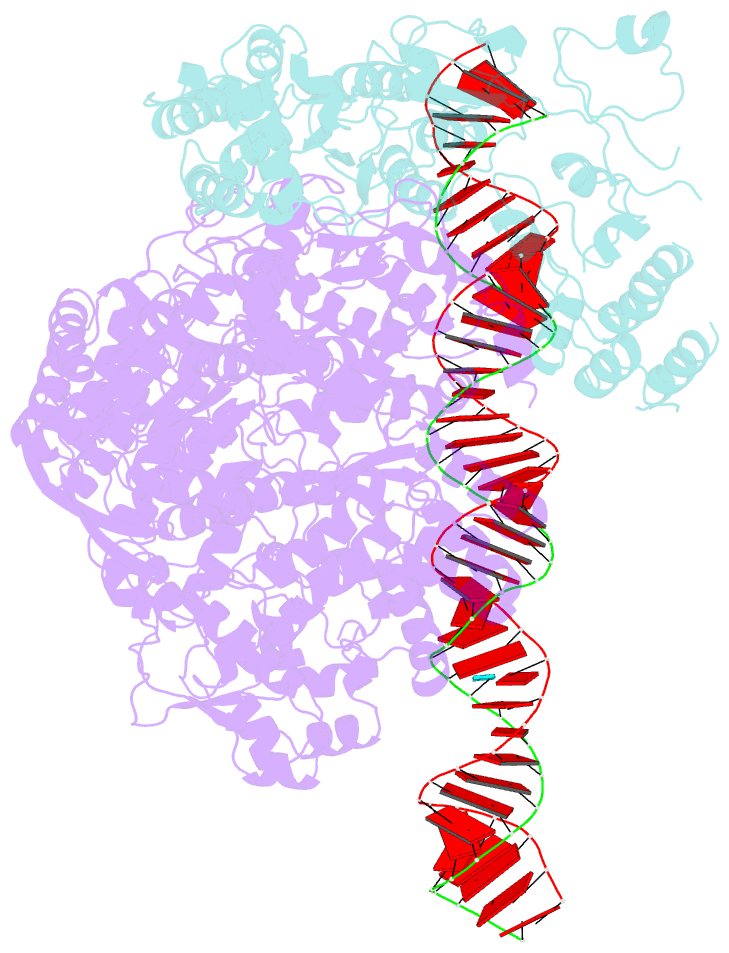
Summary information and primary citation
- PDB-id
- 5h0r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- cryo-EM (3.9 Å)
- Summary
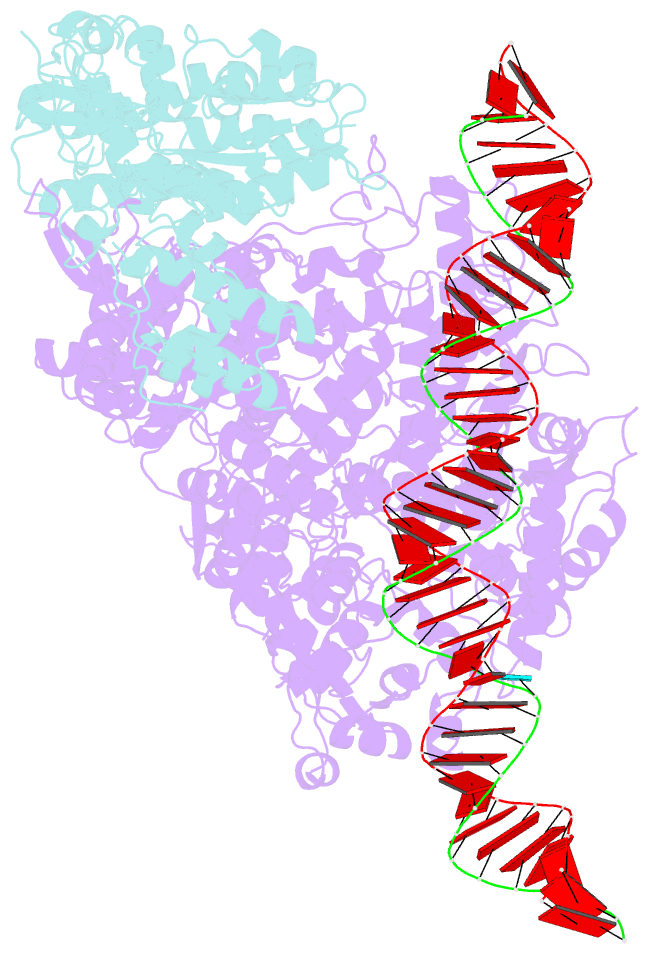
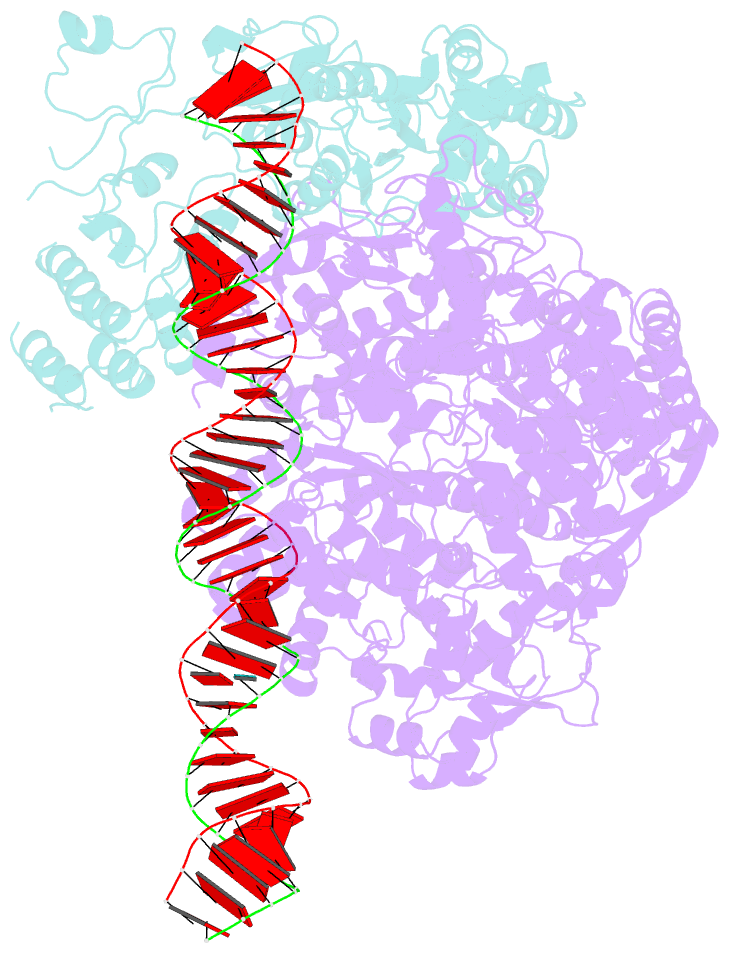
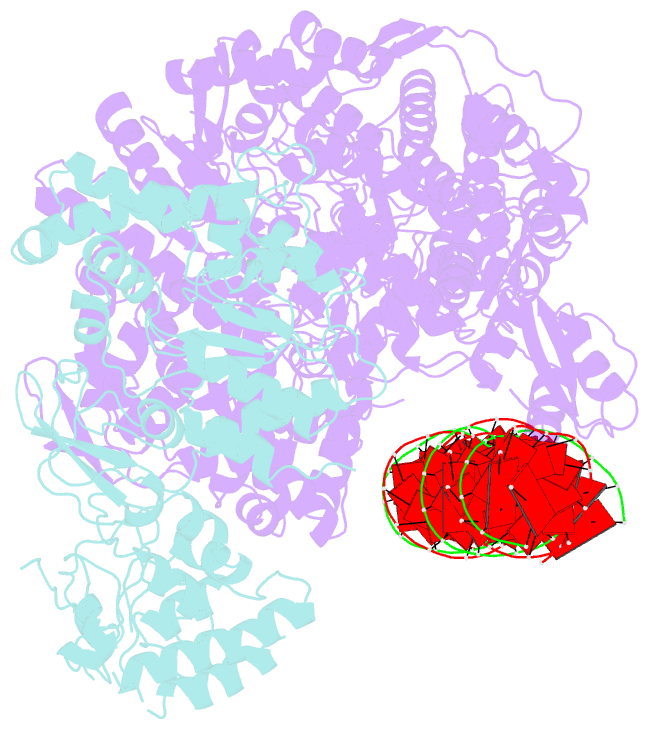
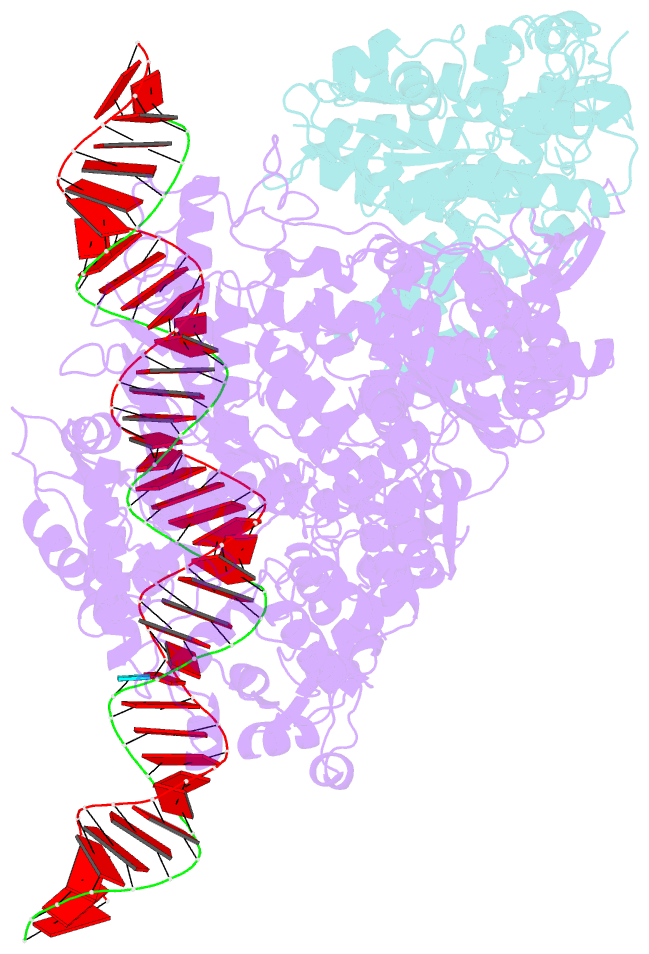
- RNA dependent RNA polymerase ,vp4,dsrna
- Reference
- Li X, Zhou N, Chen W, Zhu B, Wang X, Xu B, Wang J, Liu H, Cheng L (2017): "Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV." J. Mol. Biol., 429, 79-87. doi: 10.1016/j.jmb.2016.11.025.
- Abstract
- Single-particle cryo-electron microscopy (cryo-EM) allows the high-resolution structural determination of biological assemblies in a near-native environment. However, all high-resolution (better than 3.5Å) cryo-EM structures reported to date were obtained by using 300kV transmission electron microscopes (TEMs). We report here the structures of a cypovirus capsid of 750-Å diameter at 3.3-Å resolution and of RNA-dependent RNA polymerase (RdRp) complexes within the capsid at 3.9-Å resolution using a 200-kV TEM. The newly resolved structure revealed conformational changes of two subdomains in the RdRp. These conformational changes, which were involved in RdRp's switch from non-transcribing to transcribing mode, suggest that the RdRp may facilitate the unwinding of genomic double-stranded RNA. The possibility of 3-Å resolution structural determinations for biological assemblies of relatively small sizes using cryo-EM at 200kV was discussed.