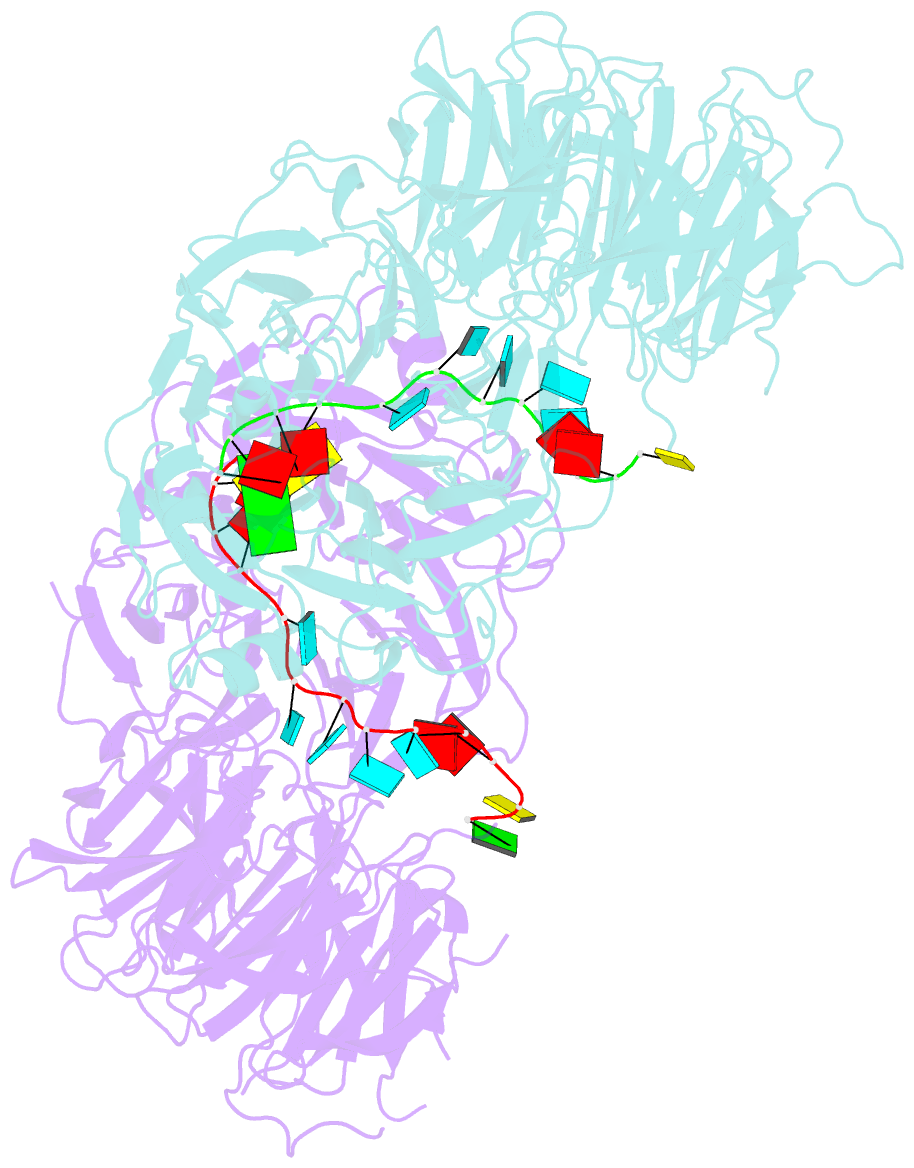
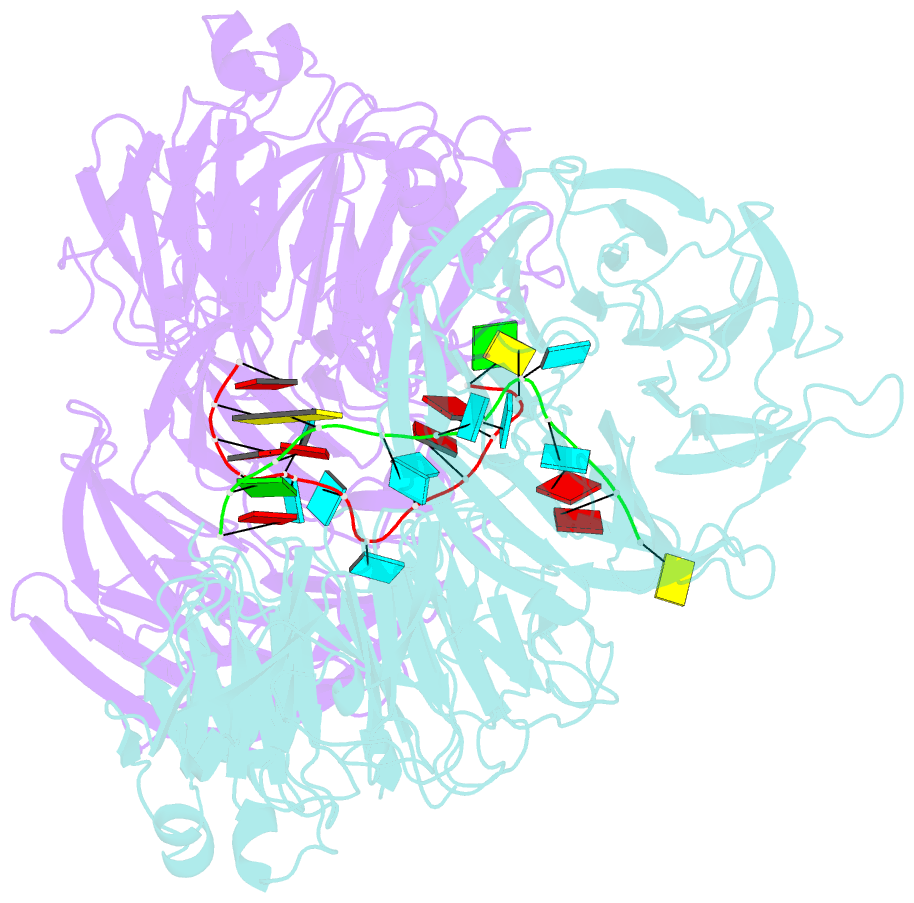
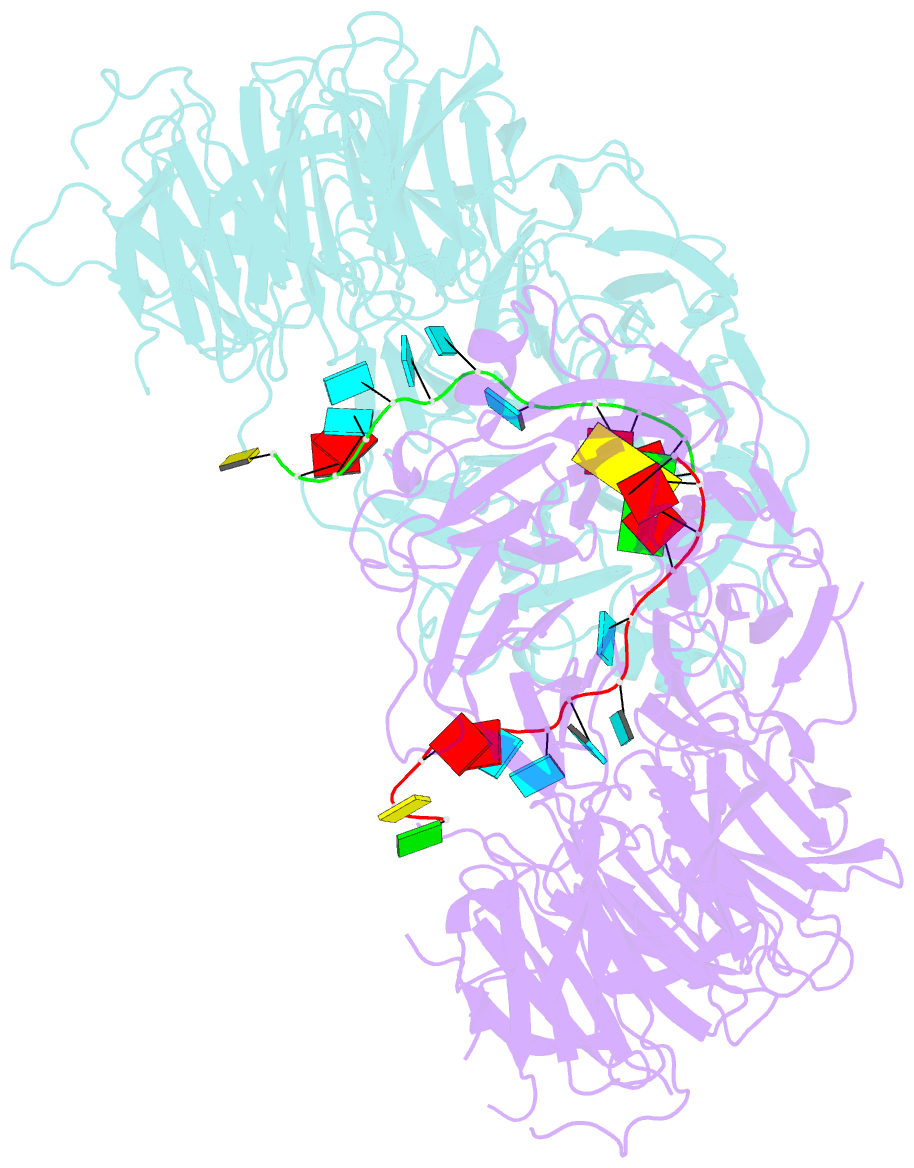
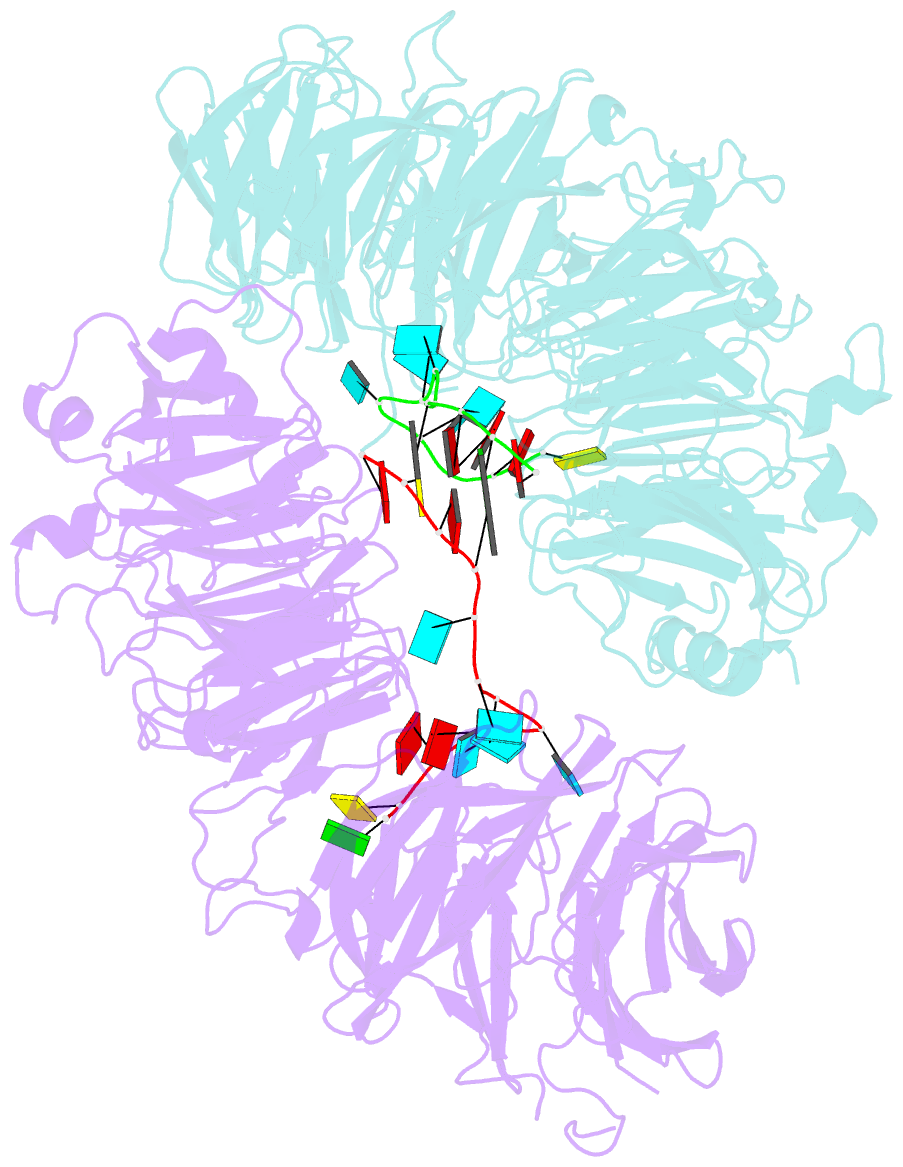
Summary information and primary citation
- PDB-id
- 5h1k; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing-RNA
- Method
- X-ray (1.9 Å)
- Summary
- Crystal structure of wd40 repeat domains of gemin5 in complex with 13-nt u4 snrna fragment
- Reference
- Jin W, Wang Y, Liu CP, Yang N, Jin M, Cong Y, Wang M, Xu RM (2016): "Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5." Genes Dev., 30, 2391-2403. doi: 10.1101/gad.291377.116.
- Abstract
- Assembly of the spliceosomal small nuclear ribonucleoparticle (snRNP) core requires the participation of the multisubunit SMN (survival of motor neuron) complex, which contains SMN and several Gemin proteins. The SMN and Gemin2 subunits directly bind Sm proteins, and Gemin5 is required for snRNP biogenesis and has been implicated in snRNA recognition. The RNA sequence required for snRNP assembly includes the Sm site and an adjacent 3' stem-loop, but a precise understanding of Gemin5's RNA-binding specificity is lacking. Here we show that the N-terminal half of Gemin5, which is composed of two juxtaposed seven-bladed WD40 repeat domains, recognizes the Sm site. The tandem WD40 repeat domains are rigidly held together to form a contiguous RNA-binding surface. RNA-contacting residues are located mostly on loops between β strands on the apical surface of the WD40 domains. Structural and biochemical analyses show that base-stacking interactions involving four aromatic residues and hydrogen bonding by a pair of arginines are crucial for specific recognition of the Sm sequence. We also show that an adenine immediately 5' to the Sm site is required for efficient binding and that Gemin5 can bind short RNA oligos in an alternative mode. Our results provide mechanistic understandings of Gemin5's snRNA-binding specificity as well as valuable insights into the molecular mechanism of RNA binding by WD40 repeat proteins in general.