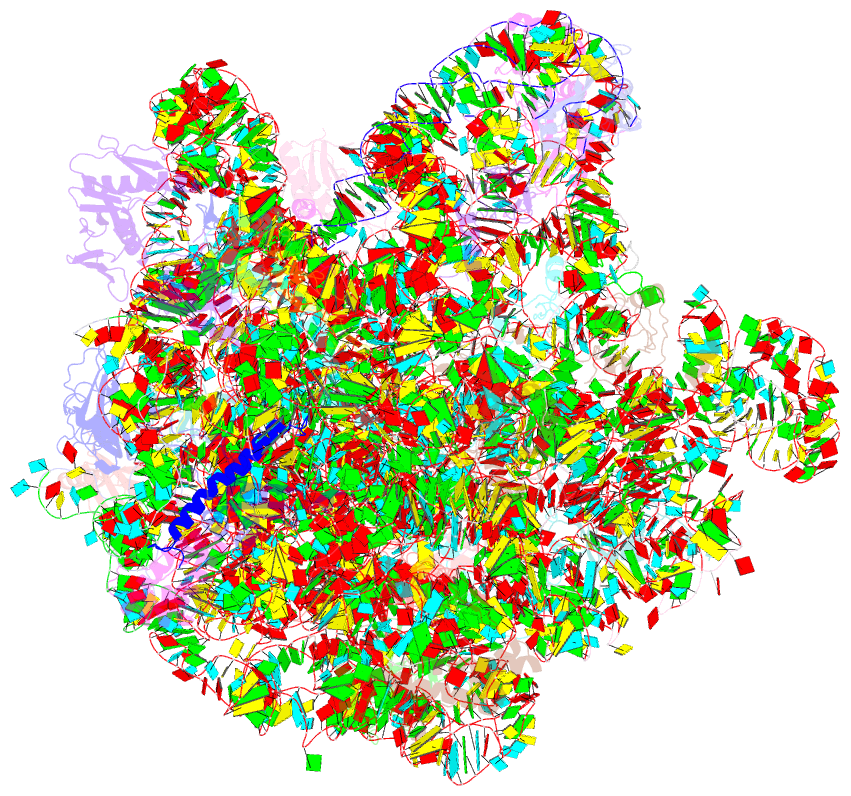
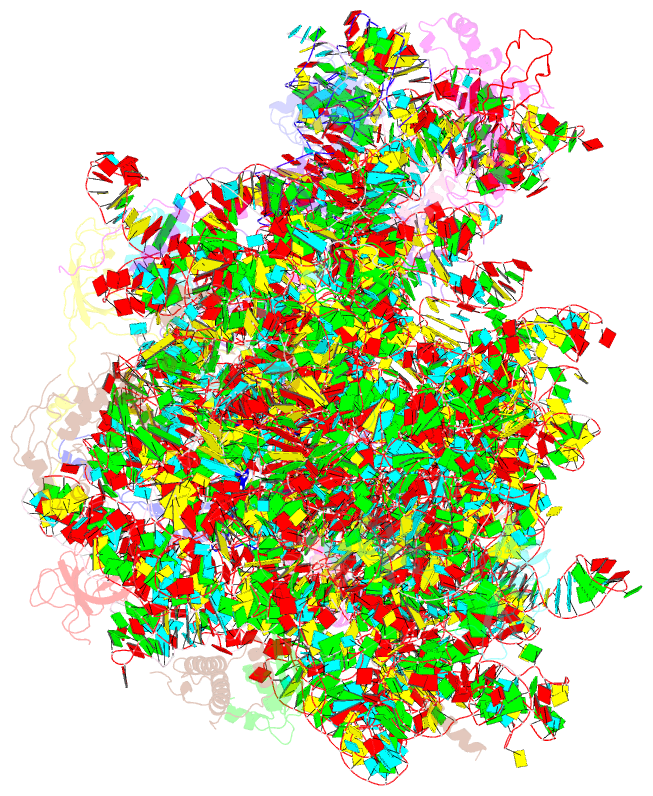
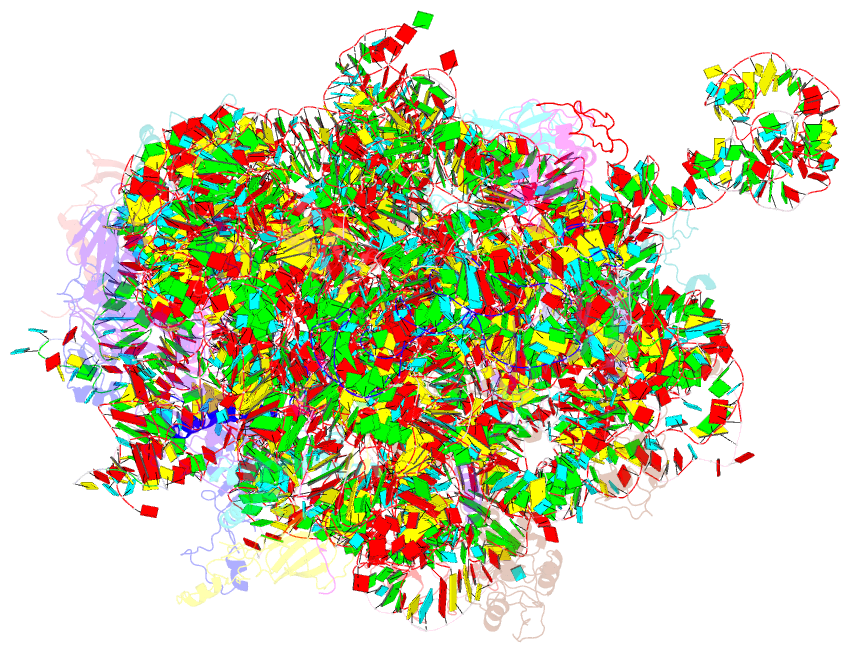
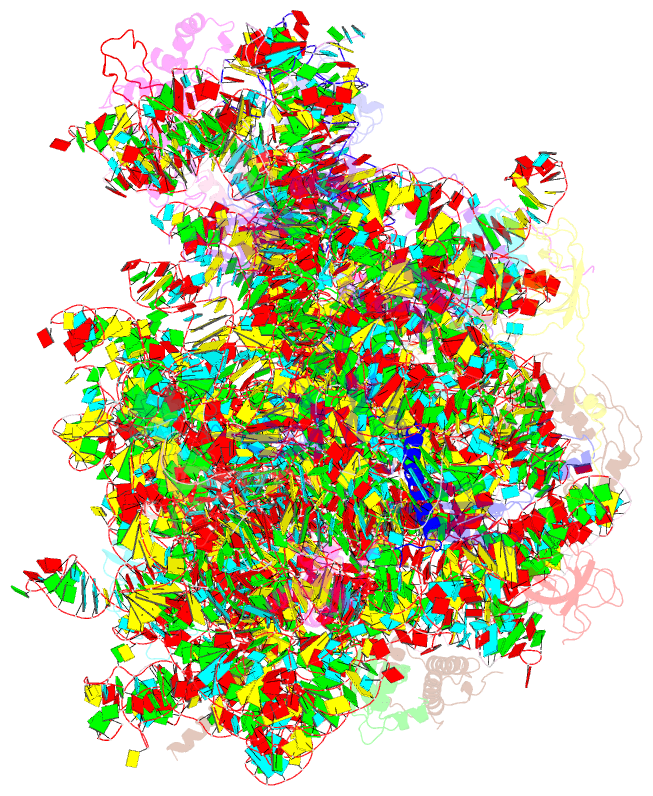
Summary information and primary citation
- PDB-id
- 5h1s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (3.5 Å)
- Summary
- Structure of the large subunit of the chloro-ribosome
- Reference
- Ahmed T, Yin Z, Bhushan S (2016): "Cryo-EM structure of the large subunit of the spinach chloroplast ribosome." Sci Rep, 6, 35793. doi: 10.1038/srep35793.
- Abstract
- Protein synthesis in the chloroplast is mediated by the chloroplast ribosome (chloro-ribosome). Overall architecture of the chloro-ribosome is considerably similar to the Escherichia coli (E. coli) ribosome but certain differences are evident. The chloro-ribosome proteins are generally larger because of the presence of chloroplast-specific extensions in their N- and C-termini. The chloro-ribosome harbours six plastid-specific ribosomal proteins (PSRPs); four in the small subunit and two in the large subunit. Deletions and insertions occur throughout the rRNA sequence of the chloro-ribosome (except for the conserved peptidyl transferase center region) but the overall length of the rRNAs do not change significantly, compared to the E. coli. Although, recent advancements in cryo-electron microscopy (cryo-EM) have provided detailed high-resolution structures of ribosomes from many different sources, a high-resolution structure of the chloro-ribosome is still lacking. Here, we present a cryo-EM structure of the large subunit of the chloro-ribosome from spinach (Spinacia oleracea) at an average resolution of 3.5 Å. High-resolution map enabled us to localize and model chloro-ribosome proteins, chloroplast-specific protein extensions, two PSRPs (PSRP5 and 6) and three rRNA molecules present in the chloro-ribosome. Although comparable to E. coli, the polypeptide tunnel and the tunnel exit site show chloroplast-specific features.