Summary information and primary citation
- PDB-id
- 5h3r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.67 Å)
- Summary
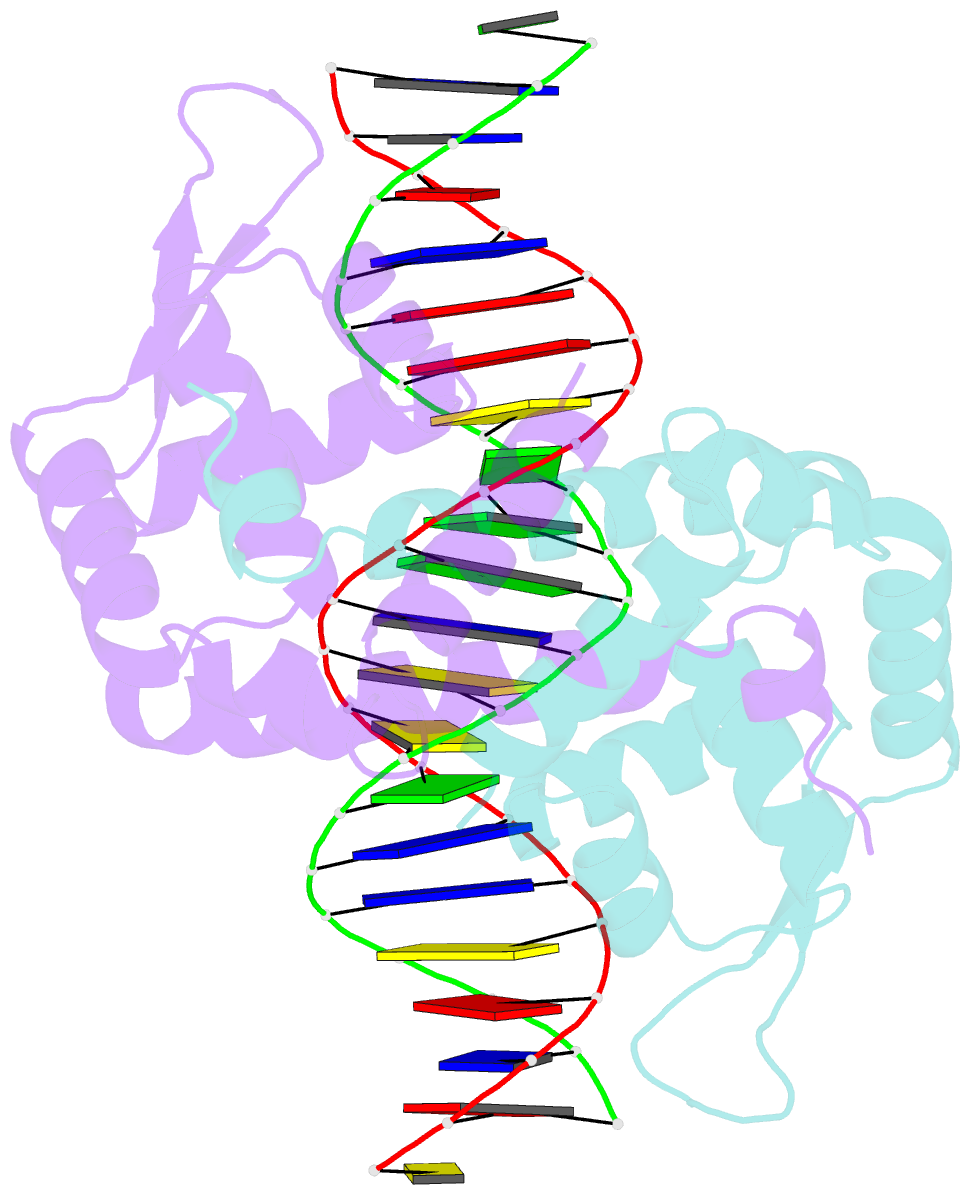
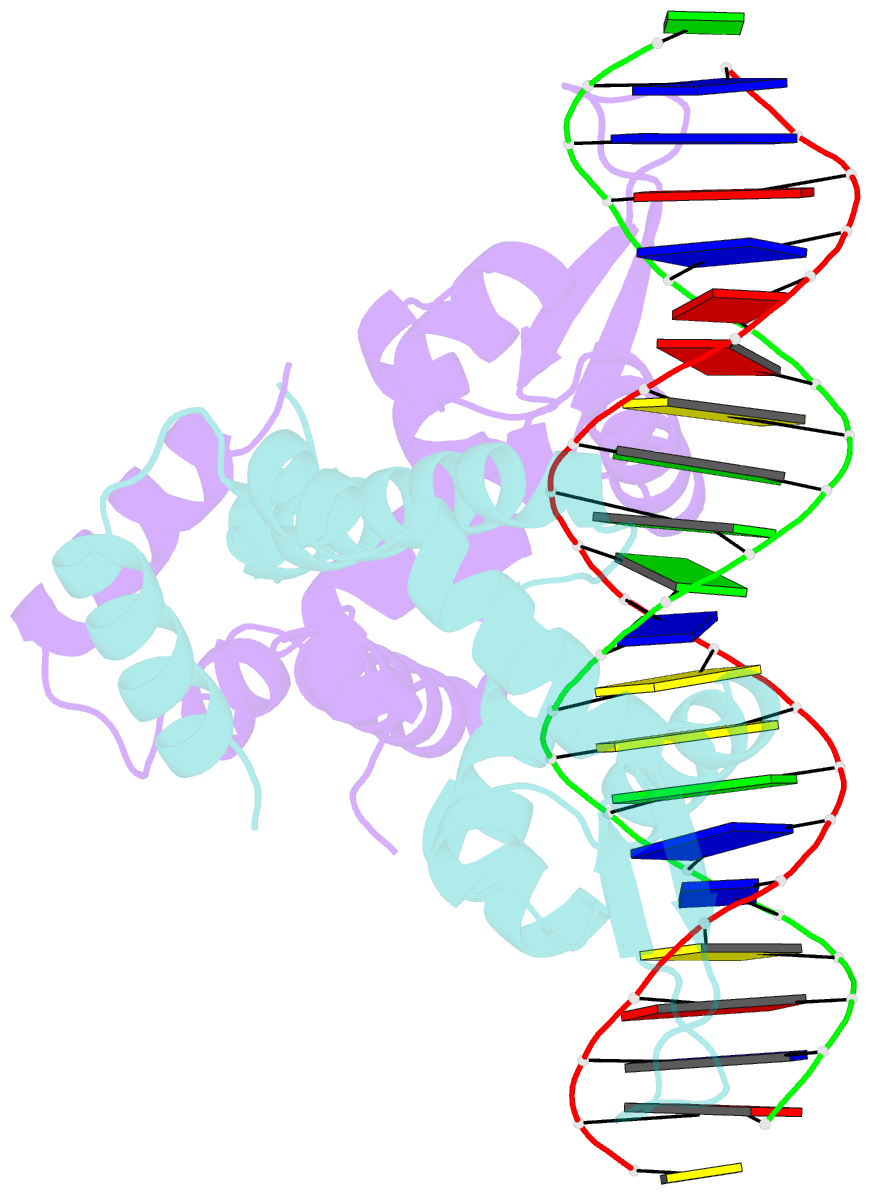
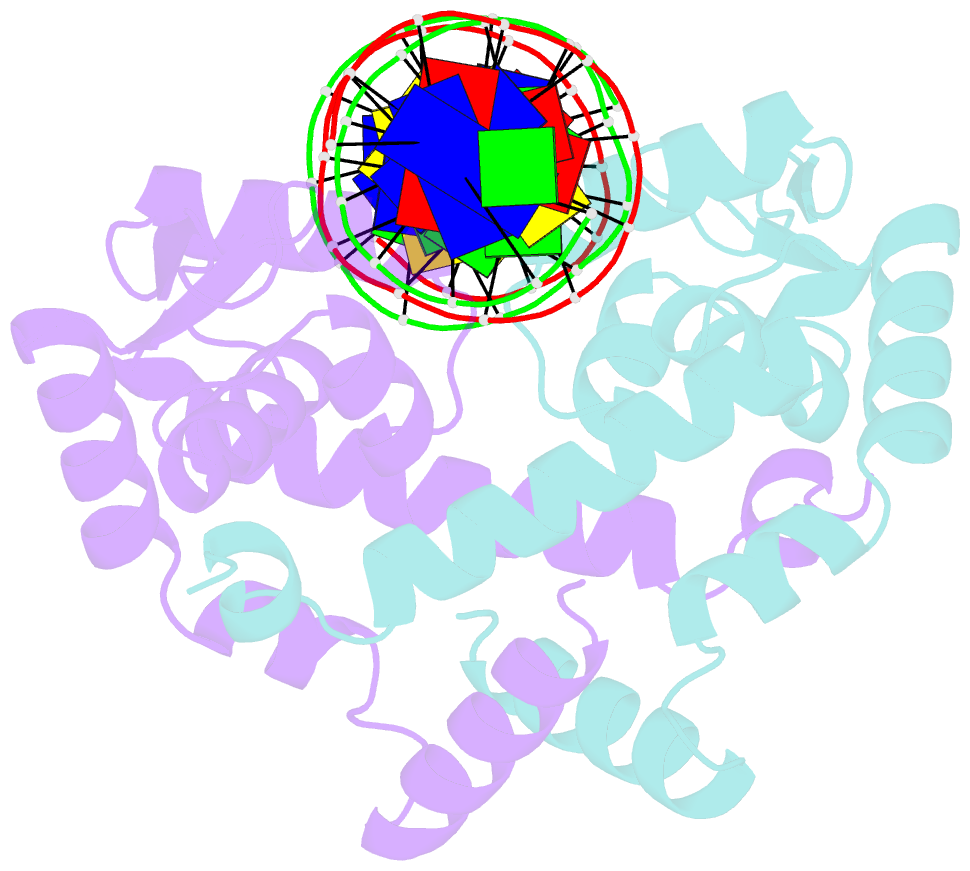
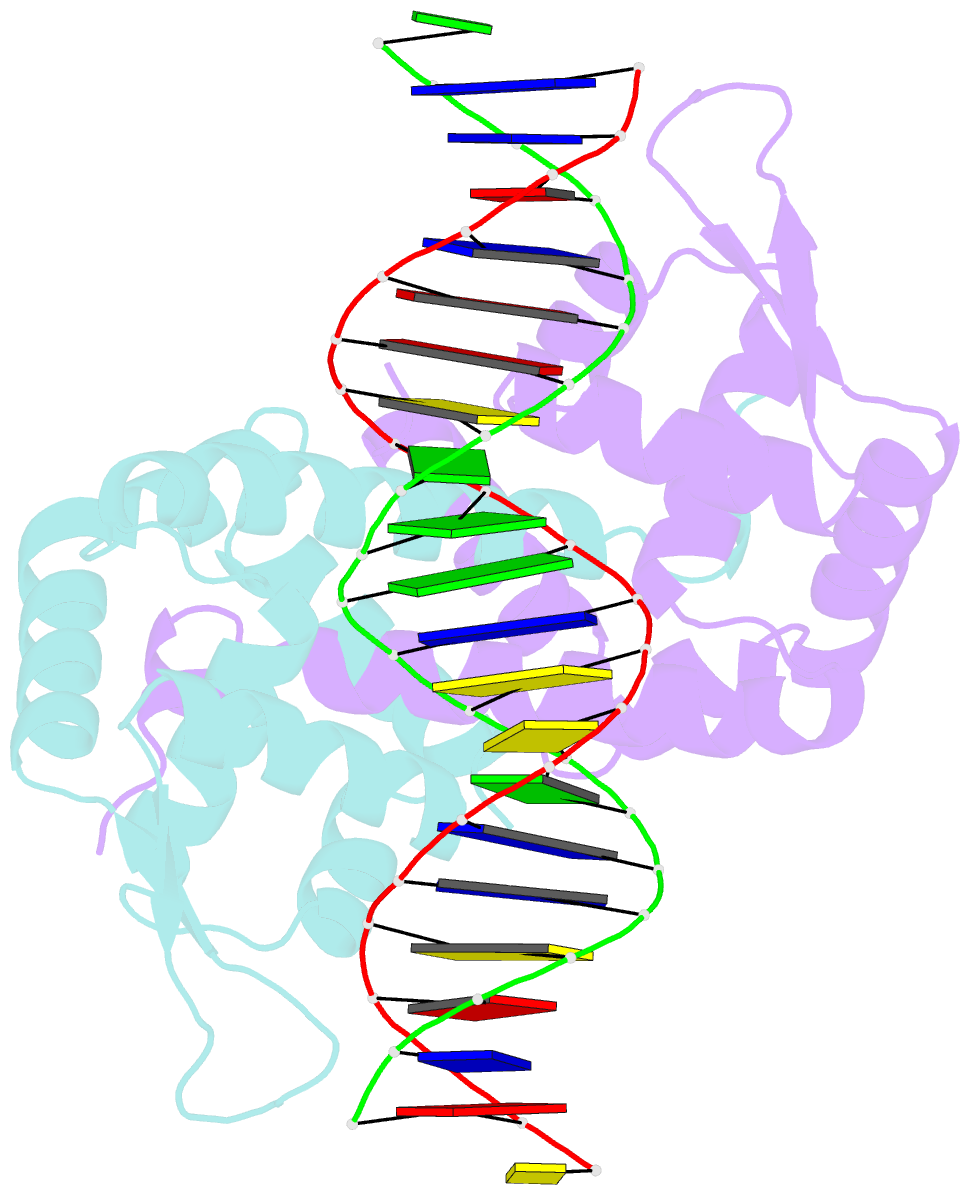
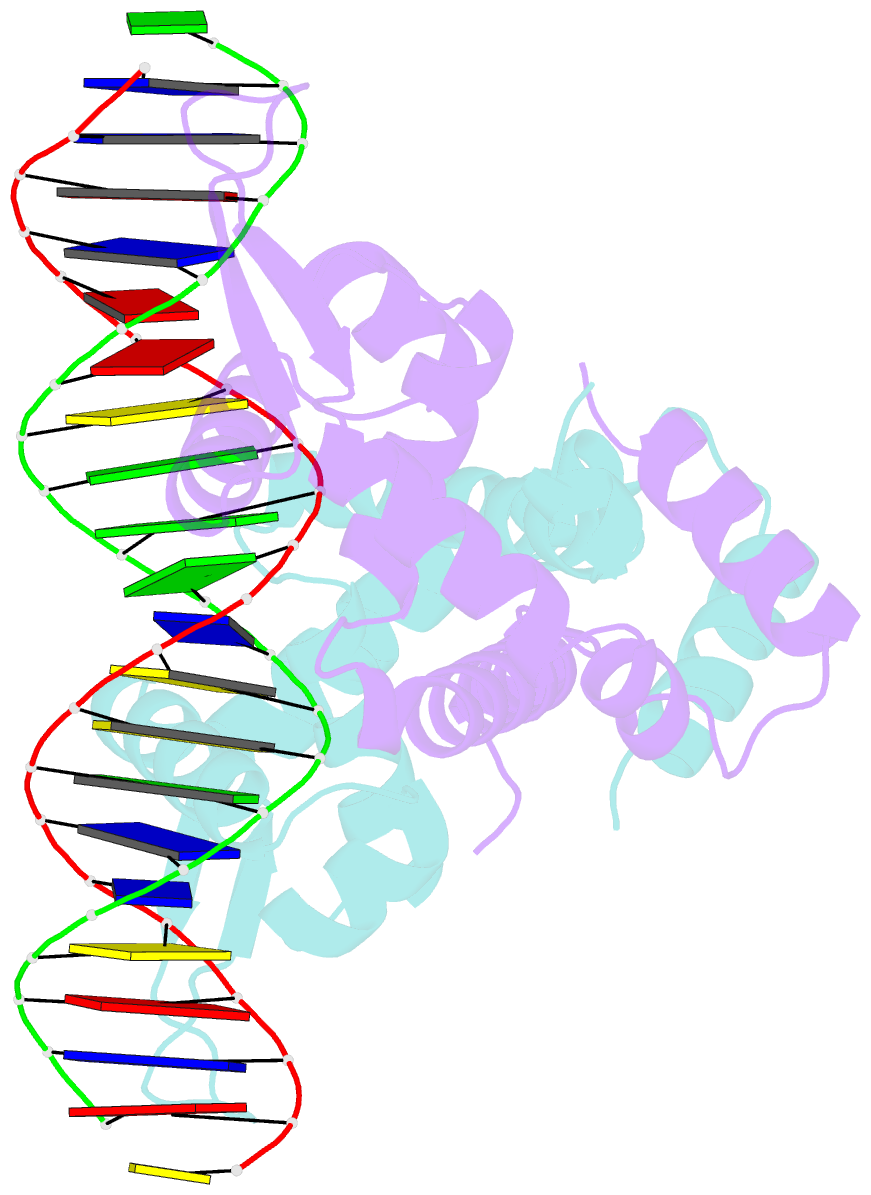
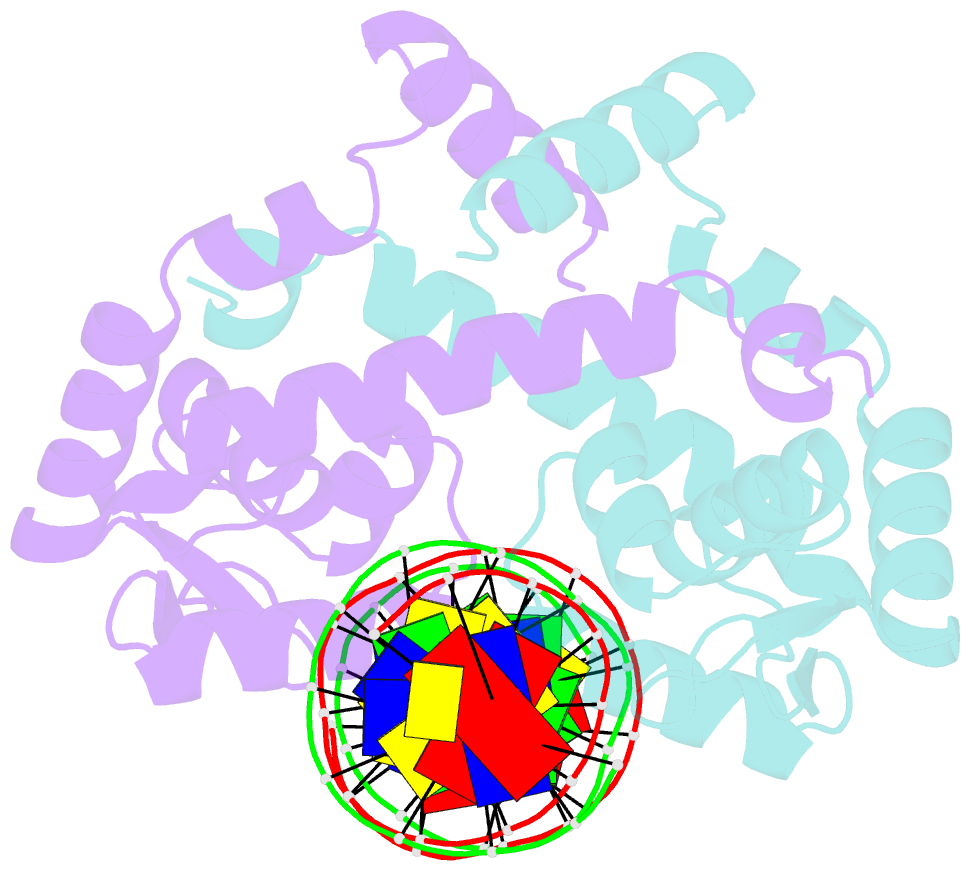
- Crystal structure of mutant marr c80s from e.coli complexed with operator DNA
- Reference
- Zhu R, Hao Z, Lou H, Song Y, Zhao J, Chen Y, Zhu J, Chen PR (2017): "Structural characterization of the DNA-binding mechanism underlying the copper(II)-sensing MarR transcriptional regulator." J. Biol. Inorg. Chem., 22, 685-693. doi: 10.1007/s00775-017-1442-7.
- Abstract
- Multiple antibiotic resistance regulator (MarR) family proteins are widely conserved transcription factors that control bacterial resistance to antibiotics, environmental stresses, as well as the regulation of virulence determinants. Escherichia coli MarR, the prototype member of this family, has recently been shown to undergo copper(II)-catalyzed inter-dimer disulfide bond formation via a unique cysteine residue (Cys80) residing in its DNA-binding domain. However, despite extensive structural characterization of the MarR family proteins, the structural mechanism for DNA binding of this copper(II)-sensing MarR factor remains elusive. Here, we report the crystal structures of DNA-bound forms of MarR, which revealed a unique, concerted generation of two new helix-loop-helix motifs that facilitated MarR's DNA binding. Structural analysis and electrophoretic mobility shift assays (EMSA) show that the flexibility of Gly116 in the center of helix α5 and the extensive hydrogen-bonding interactions at the N-terminus of helix α1 together assist the reorientation of the wHTH domains and stabilize MarR's DNA-bound conformation.