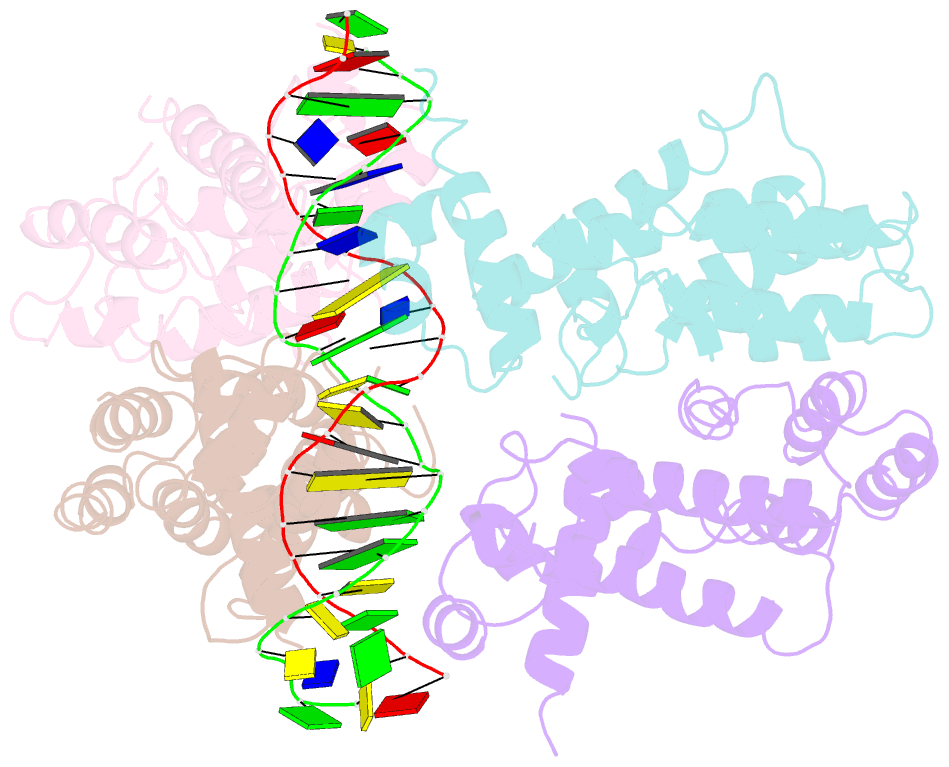
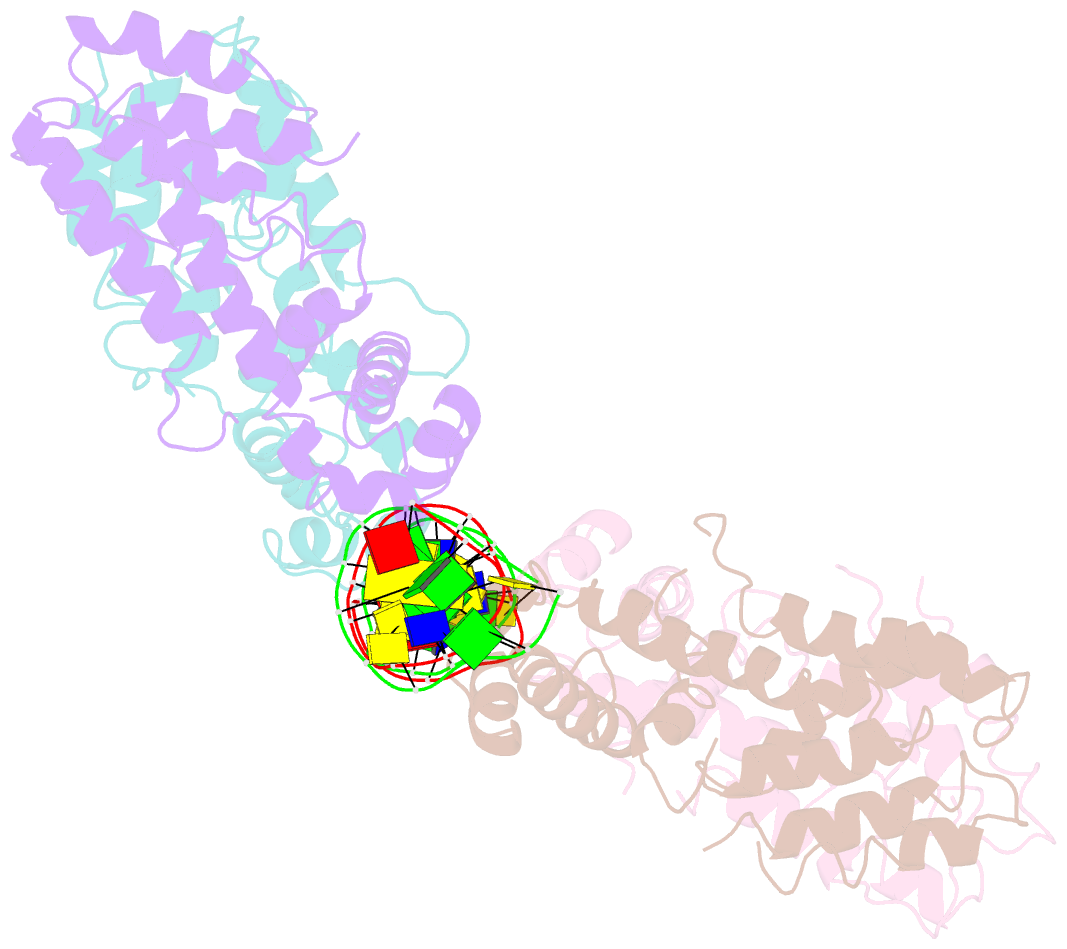
Summary information and primary citation
- PDB-id
- 5h58; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.991 Å)
- Summary
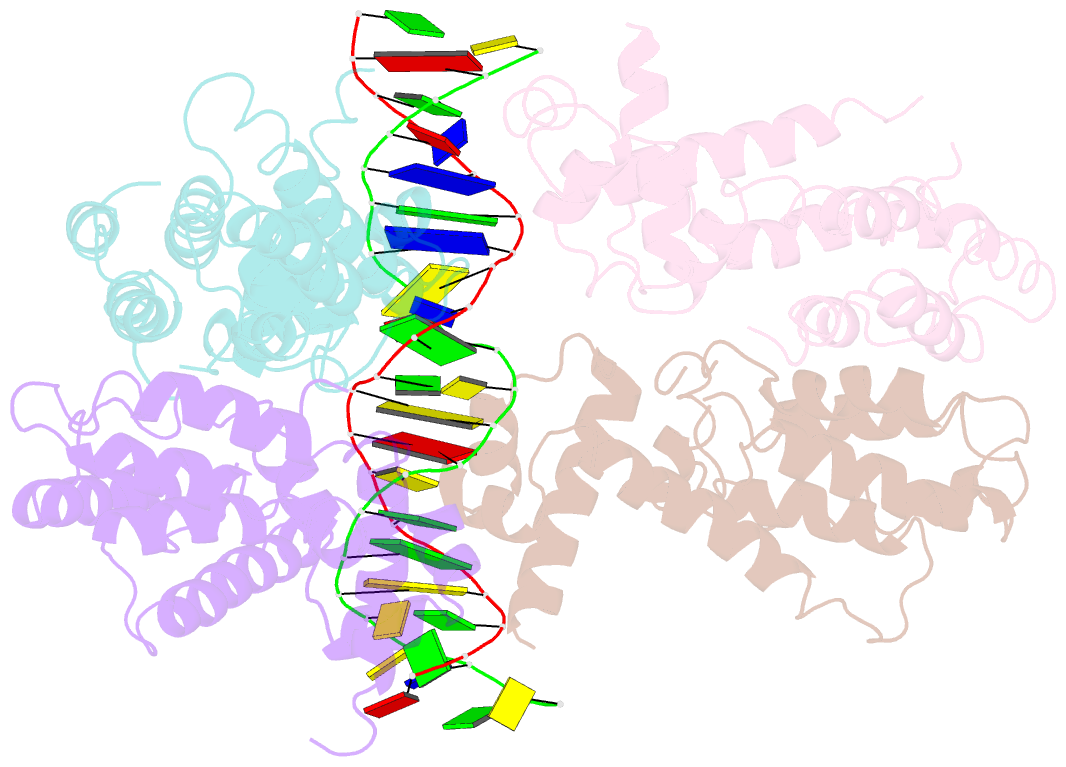
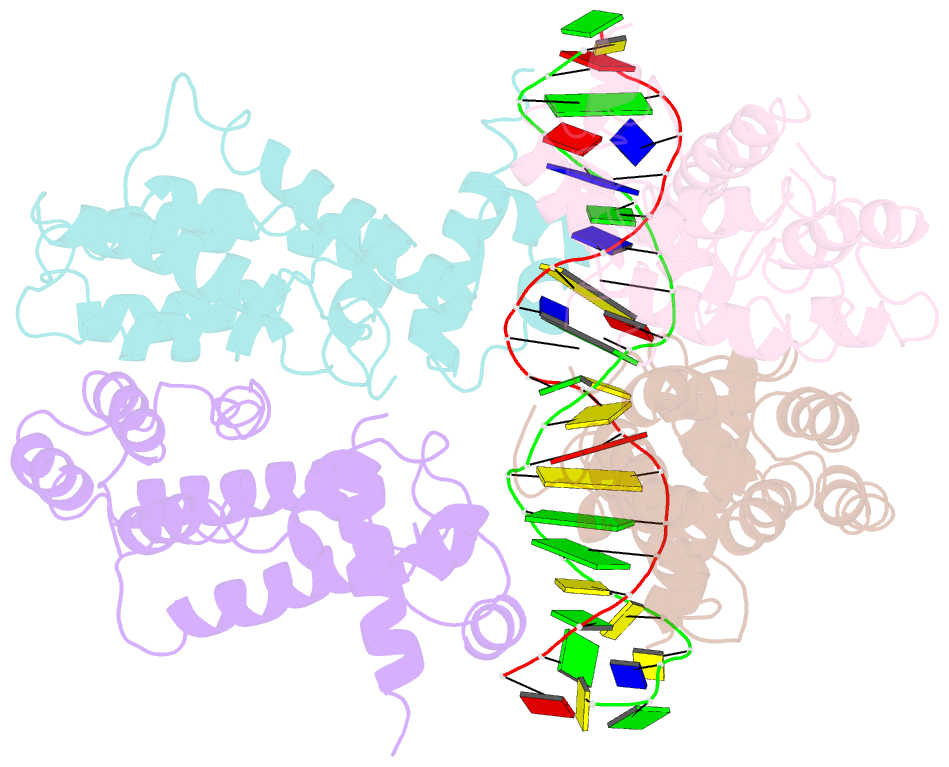
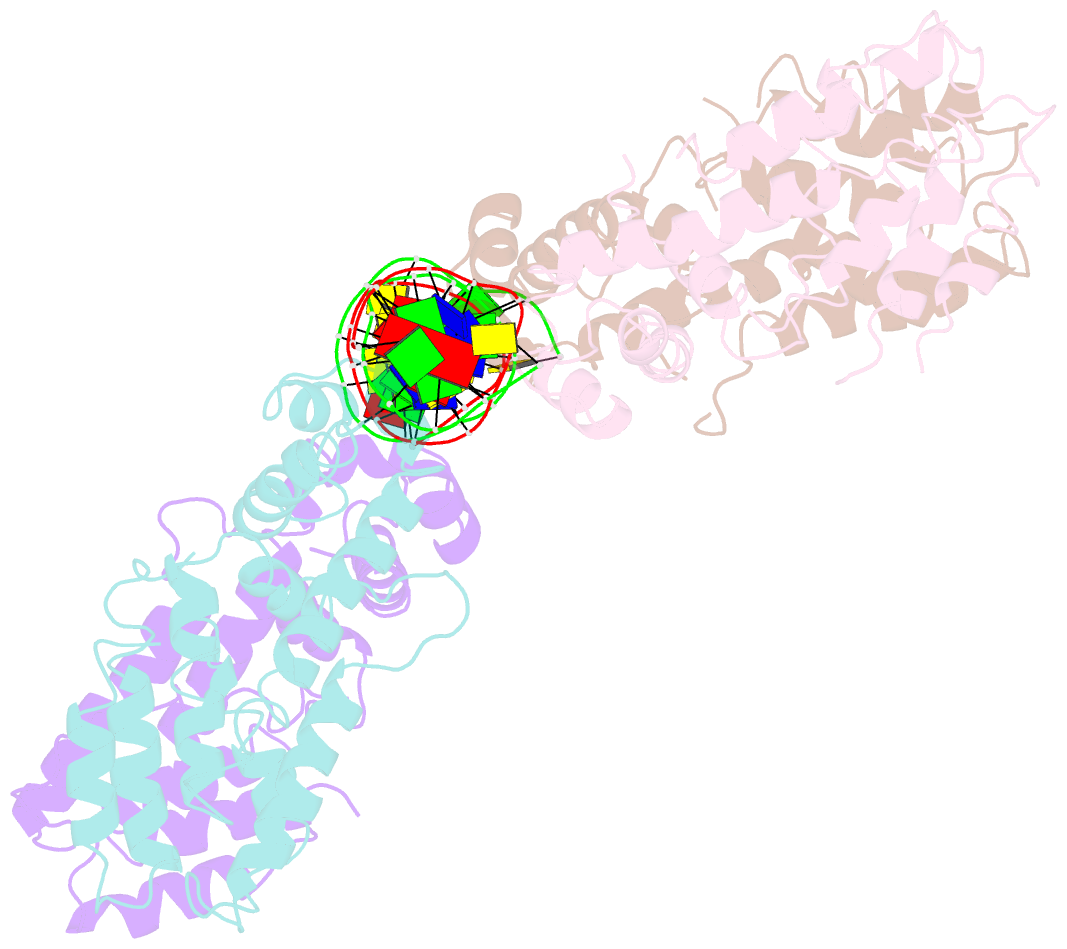
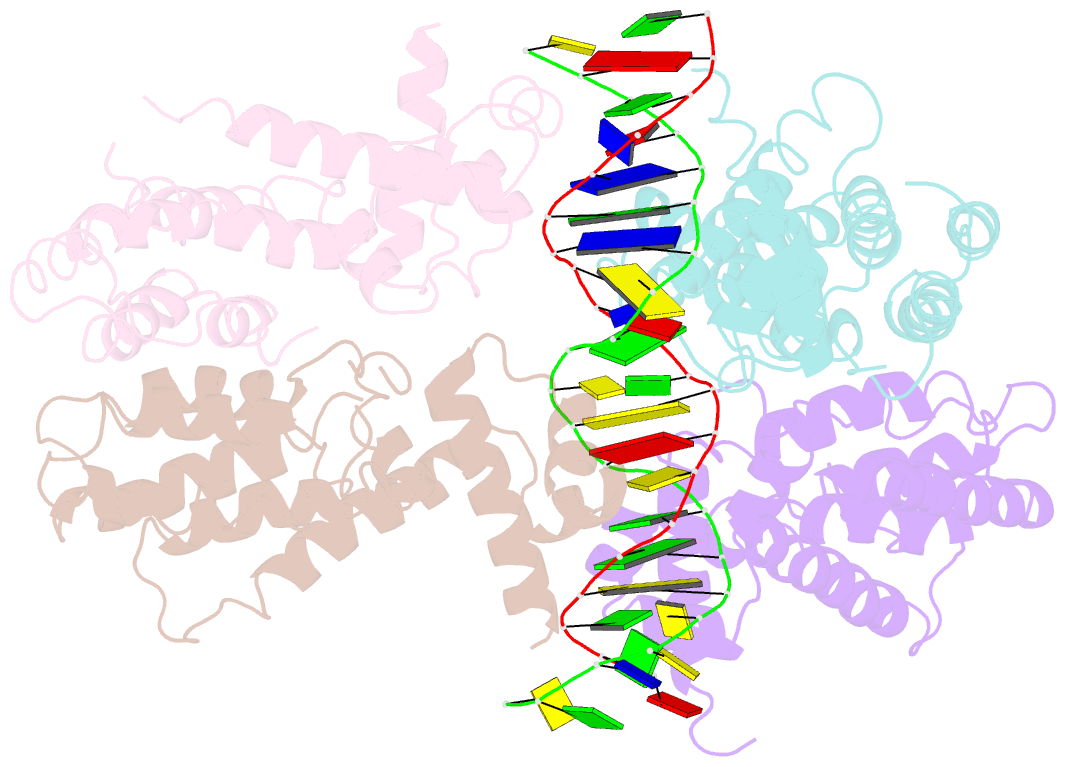
- Structural and dynamics studies of the tetr family protein, cprb from streptomyces coelicolor in complex with its biological operator sequence
- Reference
- Bhukya H, Jana AK, Sengupta N, Anand R (2017): "Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence." J. Struct. Biol., 198, 134-146. doi: 10.1016/j.jsb.2017.03.006.
- Abstract
- In Streptomycetes, tetracycline repressor family of transcription regulators (TetR-FTRs) controls various biological processes including antibiotic biosynthesis, cellular morphology and innate resistance. Here, we focus on understanding the structural basis of transcription regulation by CprB, a member of TetR-FTRs from S. coelicolor. CprB is implicated as a receptor of γ-butyrolactones, a class of quorum sensing molecules, responsible for initiating secondary metabolic pathways. In order to understand the molecular mechanism of DNA recognition, the X-ray structure of CprB in complex with its biological relevant operator sequence was solved to a resolution of 3.95Å. Furthermore, to refine and compliment the results, atomistic molecular dynamics simulations were carried out using the X-ray structure as the template. The studies reveal that CprB binds to DNA as dimer of dimers with this mode of interaction results in minimal distortion in the DNA, enabling these proteins to recognize multiple sequences with varying affinity. Another crucial finding from our simulation results was that the positively charged N-terminal arm of CprB brings extra stability to the protein-DNA complex by interacting with the minor-groove of the DNA and anchoring itself to the phosphate backbone. Corroborating electrophoretic mobility shift assay and fluorescence anisotropy experiments showed that the mutant ΔN6-CprB exhibited about 7-8 fold reduced DNA binding. Comparison with other TetR-FTRs reveals that this strategy is also employed by over 25% of TetR-FTRs, where N-terminal anchoring mechanism is used to enhance selectivity for a particular DNA sequence.