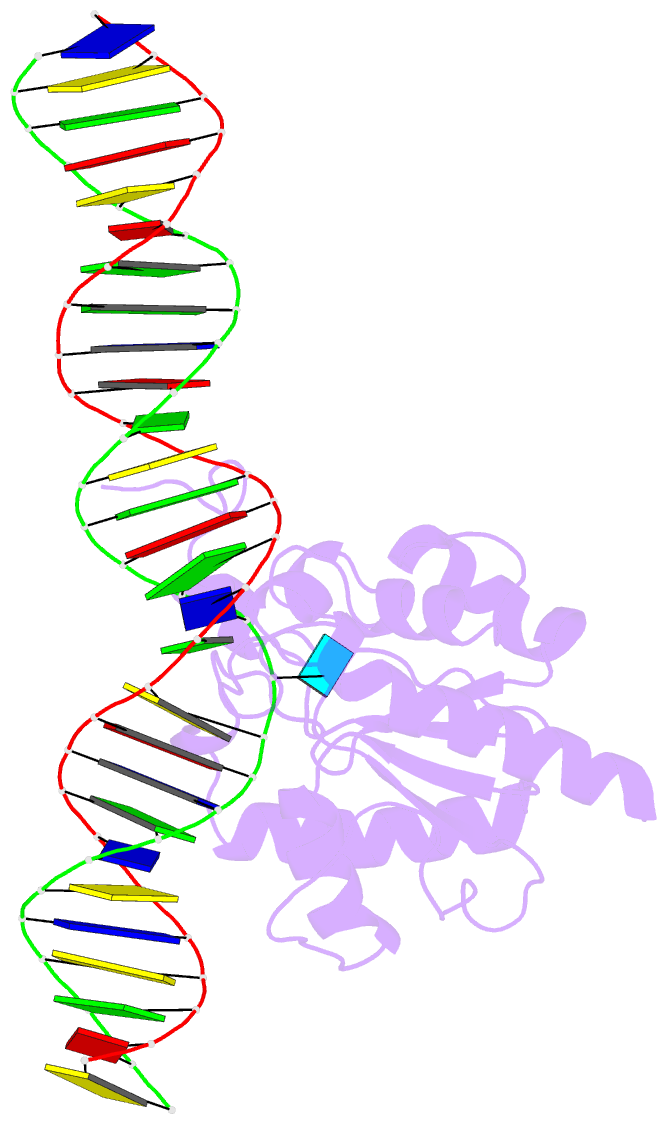
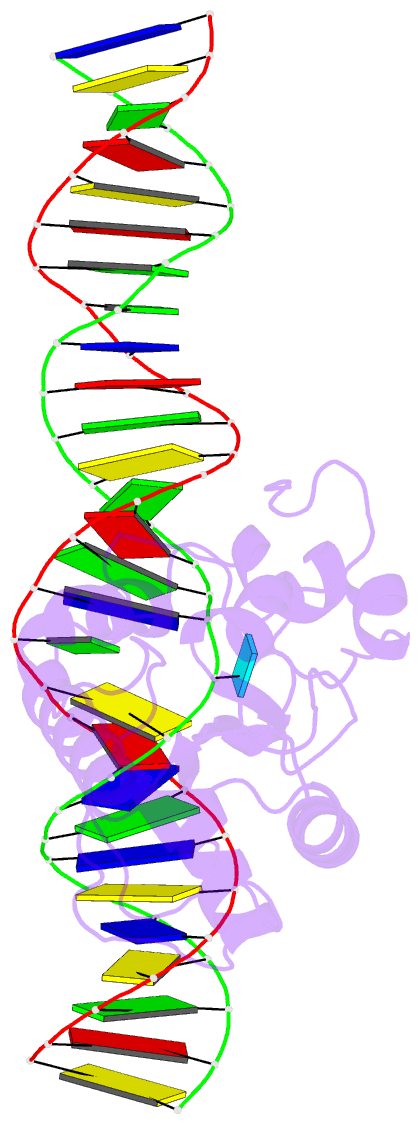
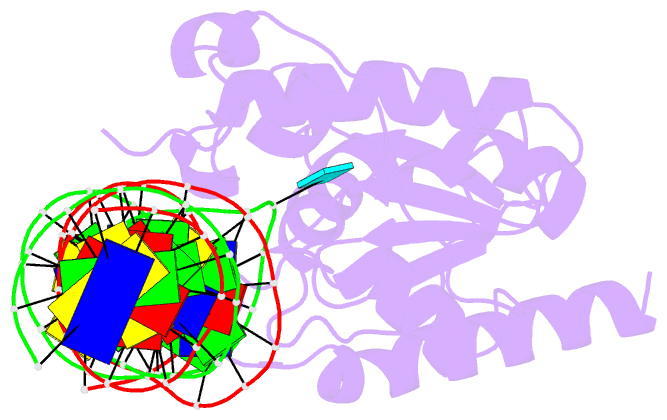
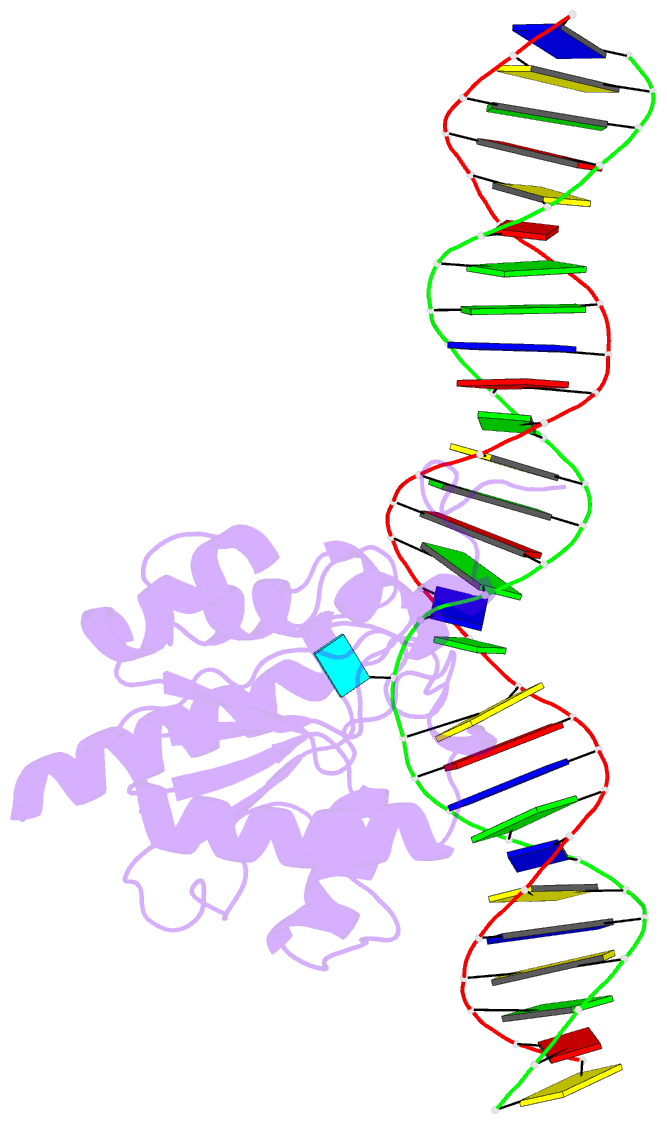
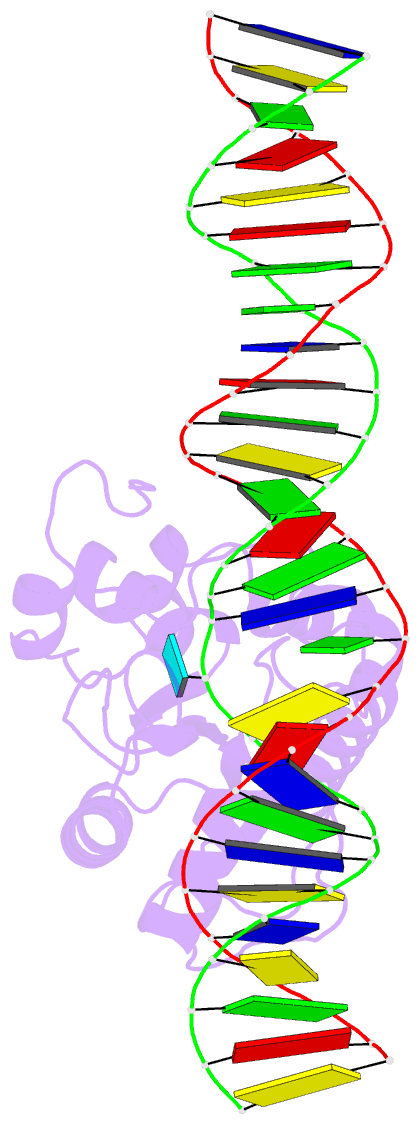
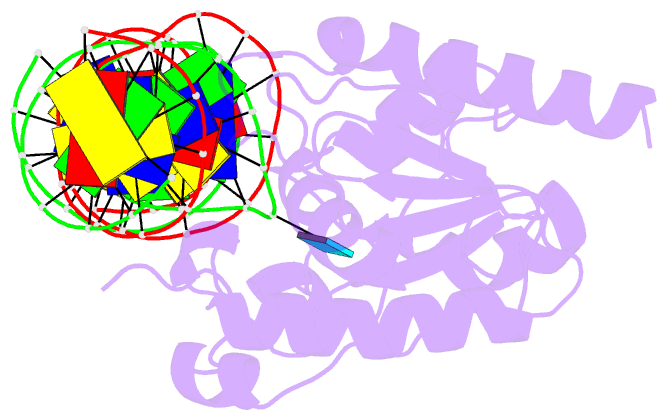
Summary information and primary citation
- PDB-id
- 5hf7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.54 Å)
- Summary
- Tdg enzyme-substrate complex
- Reference
- Coey CT, Malik SS, Pidugu LS, Varney KM, Pozharski E, Drohat AC (2016): "Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues." Nucleic Acids Res., 44, 10248-10258. doi: 10.1093/nar/gkw768.
- Abstract
- Thymine DNA Glycosylase (TDG) is a base excision repair enzyme functioning in DNA repair and epigenetic regulation. TDG removes thymine from mutagenic G·T mispairs arising from deamination of 5-methylcytosine (mC), and it processes other deamination-derived lesions including uracil (U). Essential for DNA demethylation, TDG excises 5-formylcytosine and 5-carboxylcytosine, derivatives of mC generated by Tet (ten-eleven translocation) enzymes. Here, we report structural and functional studies of TDG82-308, a new construct containing 29 more N-terminal residues than TDG111-308, the construct used for previous structures of DNA-bound TDG. Crystal structures and NMR experiments demonstrate that most of these N-terminal residues are disordered, for substrate- or product-bound TDG82-308 Nevertheless, G·T substrate affinity and glycosylase activity of TDG82-308 greatly exceeds that of TDG111-308 and is equivalent to full-length TDG. We report the first high-resolution structures of TDG in an enzyme-substrate complex, for G·U bound to TDG82-308 (1.54 Å) and TDG111-308 (1.71 Å), revealing new enzyme-substrate contacts, direct and water-mediated. We also report a structure of the TDG82-308 product complex (1.70 Å). TDG82-308 forms unique enzyme-DNA interactions, supporting its value for structure-function studies. The results advance understanding of how TDG recognizes and removes modified bases from DNA, particularly those resulting from deamination.