Summary information and primary citation
- PDB-id
- 5hl7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- X-ray (3.55 Å)
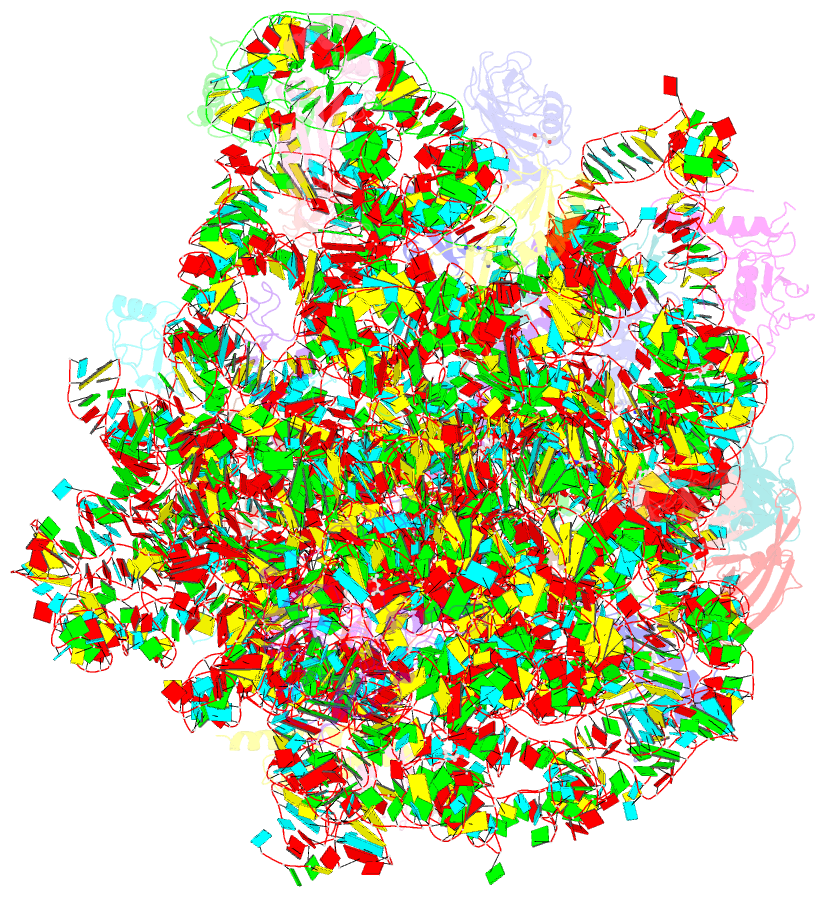
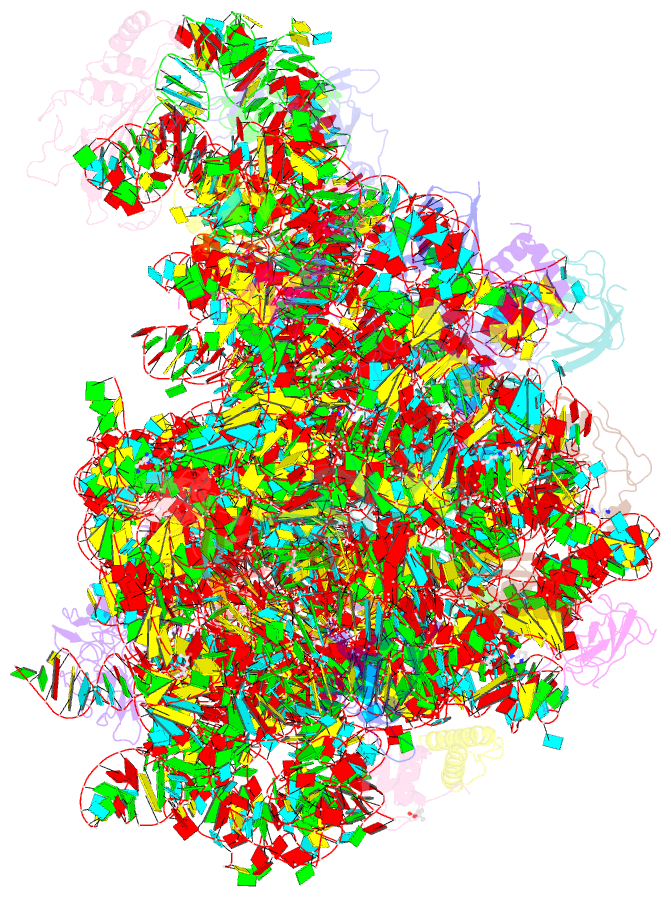
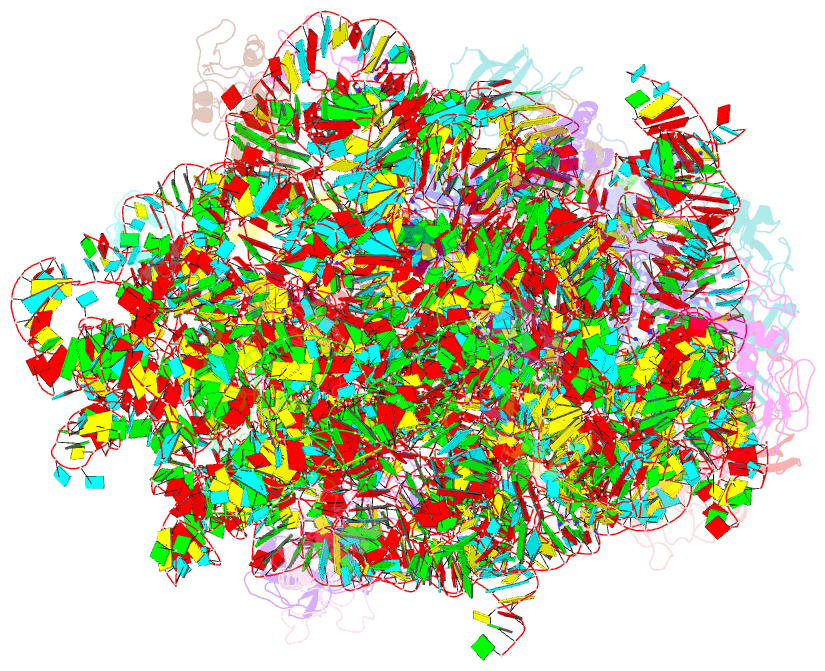
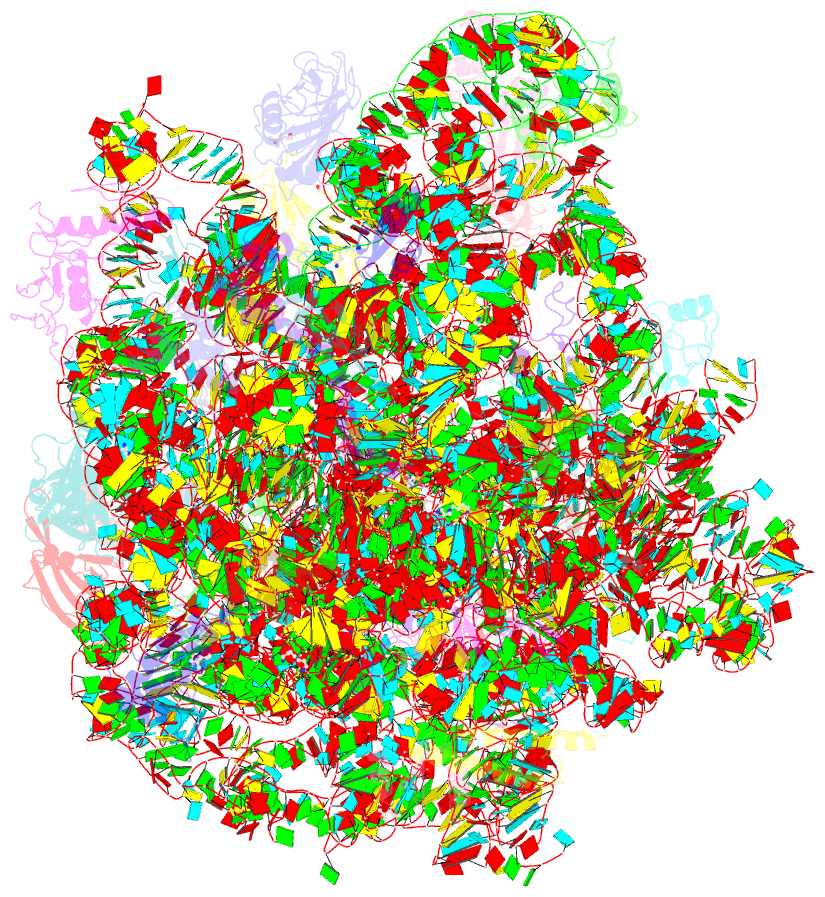
- Summary
- The crystal structure of the large ribosomal subunit of staphylococcus aureus in complex with lefamulin
- Reference
- Eyal Z, Matzov D, Krupkin M, Paukner S, Riedl R, Rozenberg H, Zimmerman E, Bashan A, Yonath A (2016): "A novel pleuromutilin antibacterial compound, its binding mode and selectivity mechanism." Sci Rep, 6, 39004. doi: 10.1038/srep39004.
- Abstract
- The increasing appearance of pathogenic bacteria with antibiotic resistance is a global threat. Consequently, clinically available potent antibiotics that are active against multidrug resistant pathogens are becoming exceedingly scarce. Ribosomes are a main target for antibiotics, and hence are an objective for novel drug development. Lefamulin, a semi-synthetic pleuromutilin compound highly active against multi-resistant pathogens, is a promising antibiotic currently in phase III trials for the treatment of community-acquired bacterial pneumonia in adults. The crystal structure of the Staphylococcus aureus large ribosomal subunit in complex with lefamulin reveals its protein synthesis inhibition mechanism and the rationale for its potency. In addition, analysis of the bacterial and eukaryotes ribosome structures around the pleuromutilin binding pocket has elucidated the key for the drug's selectivity.