Summary information and primary citation
- PDB-id
- 5hlf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-inhibitor-DNA
- Method
- X-ray (2.95 Å)
- Summary
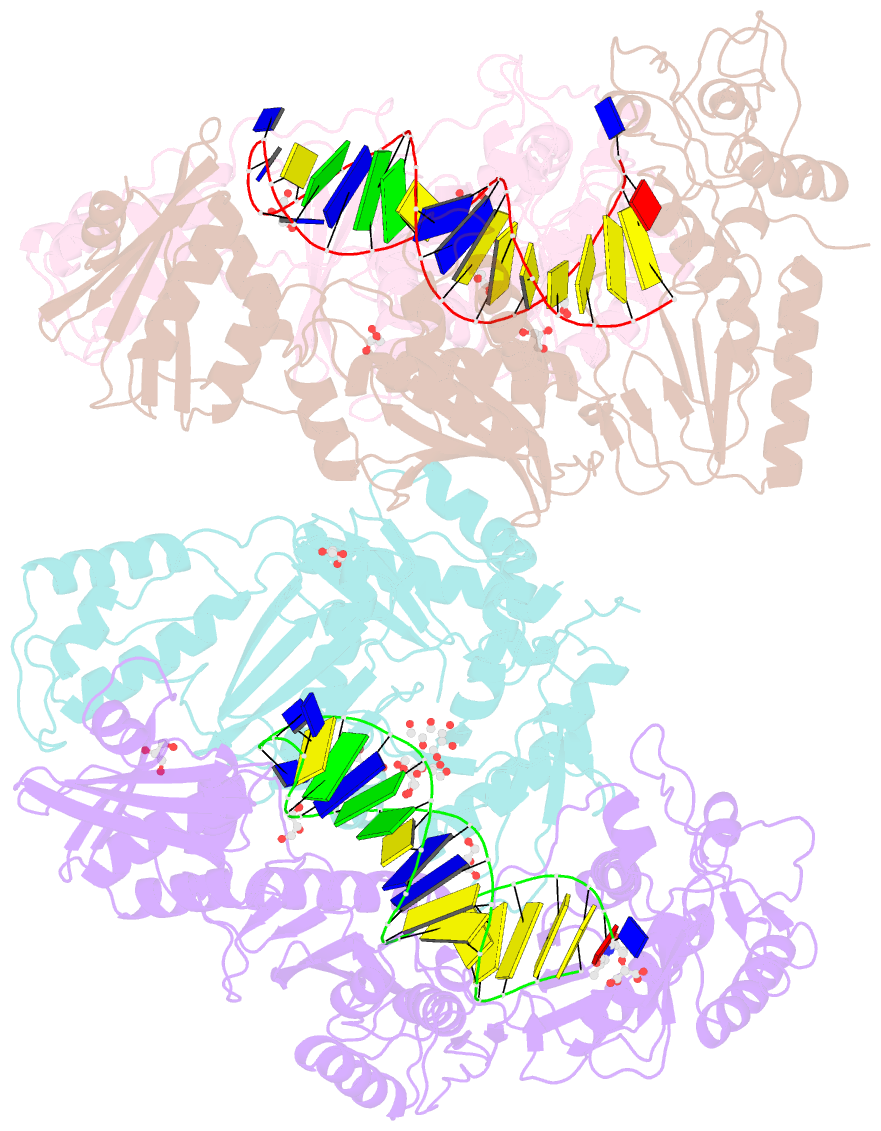
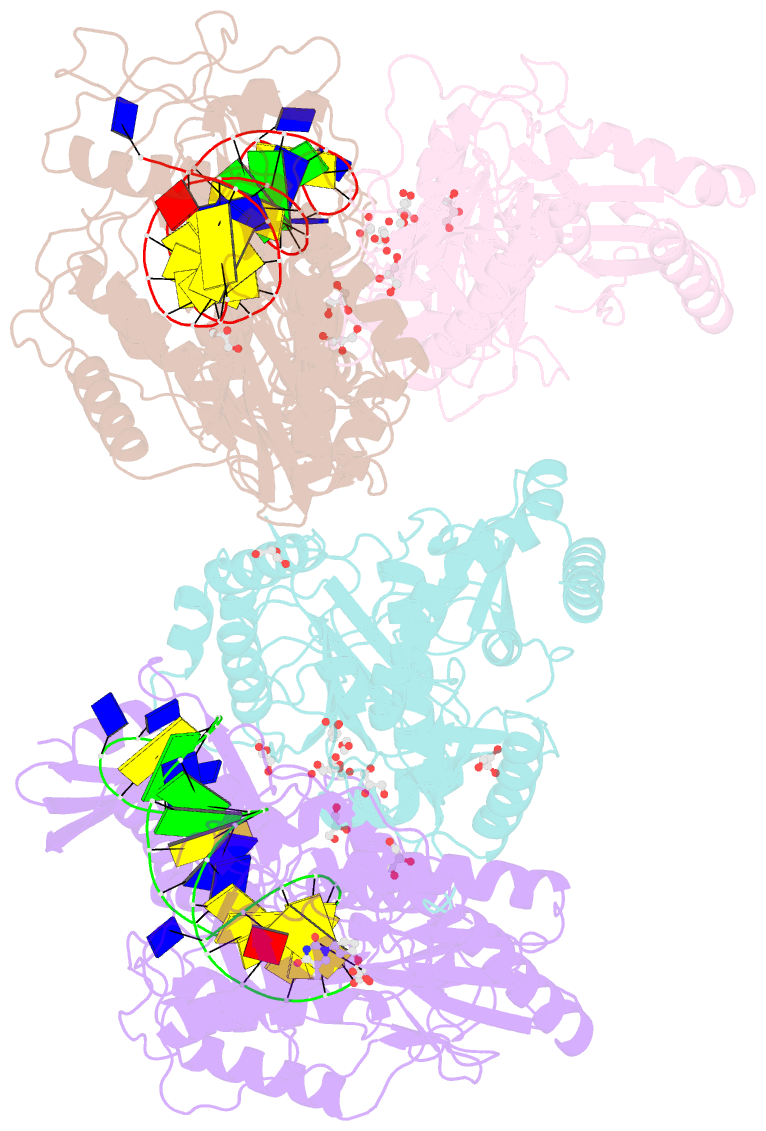
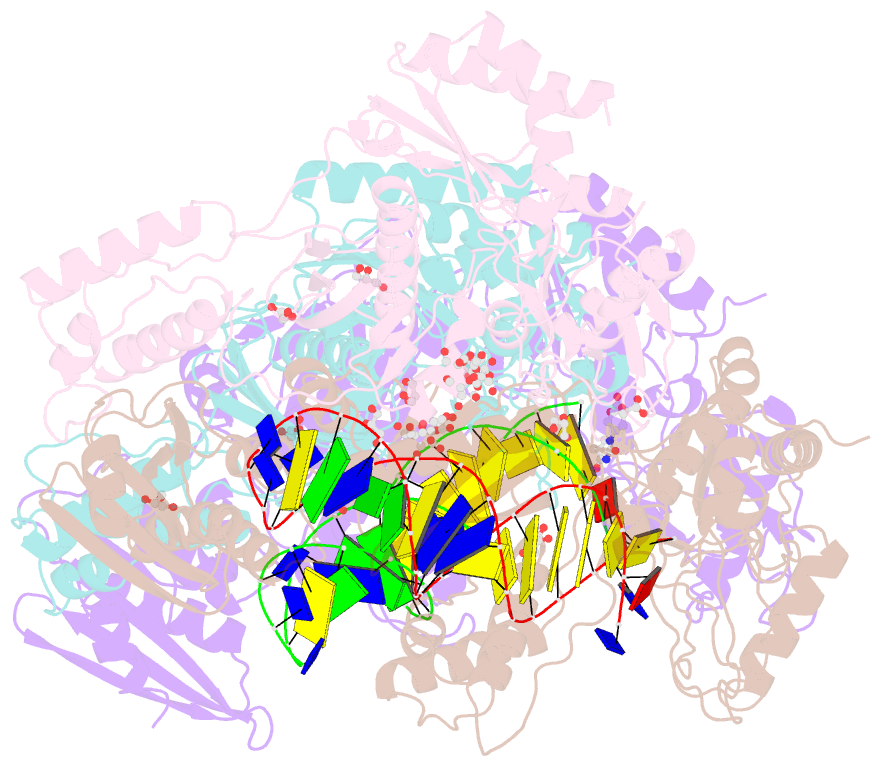
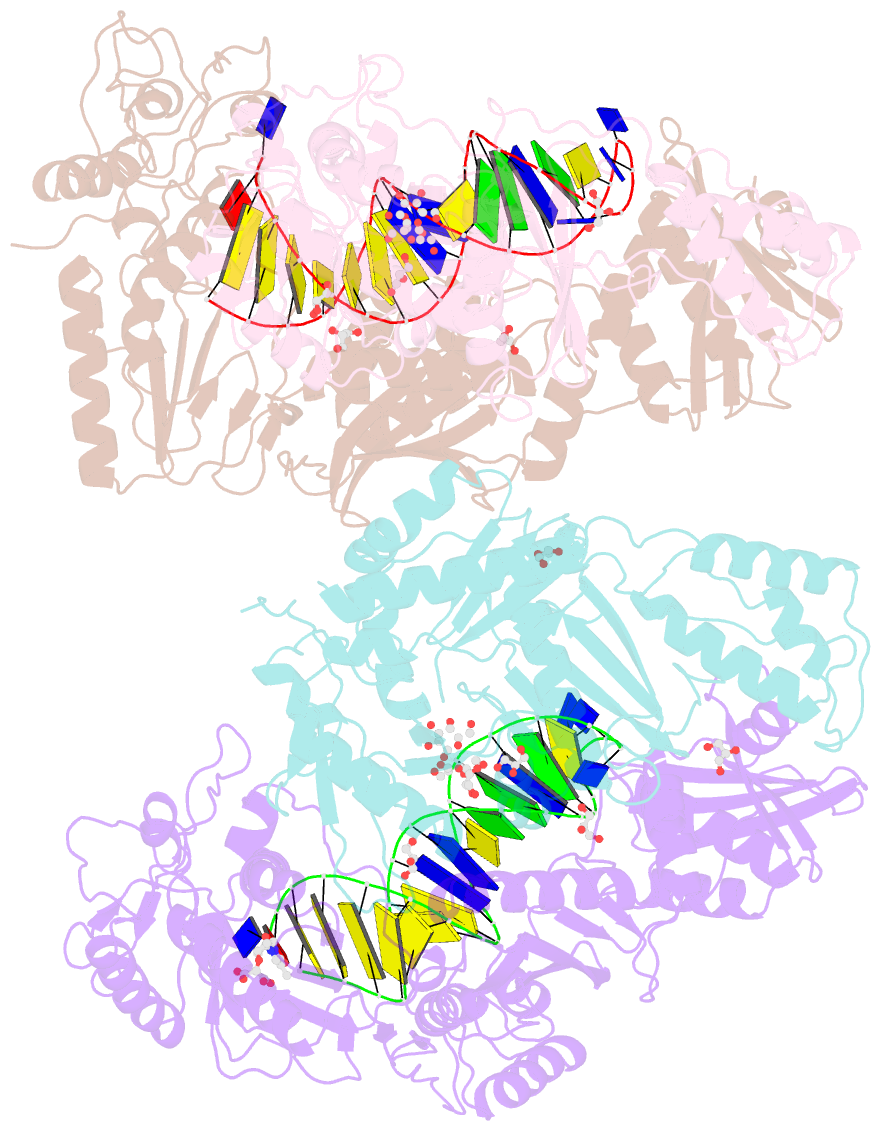
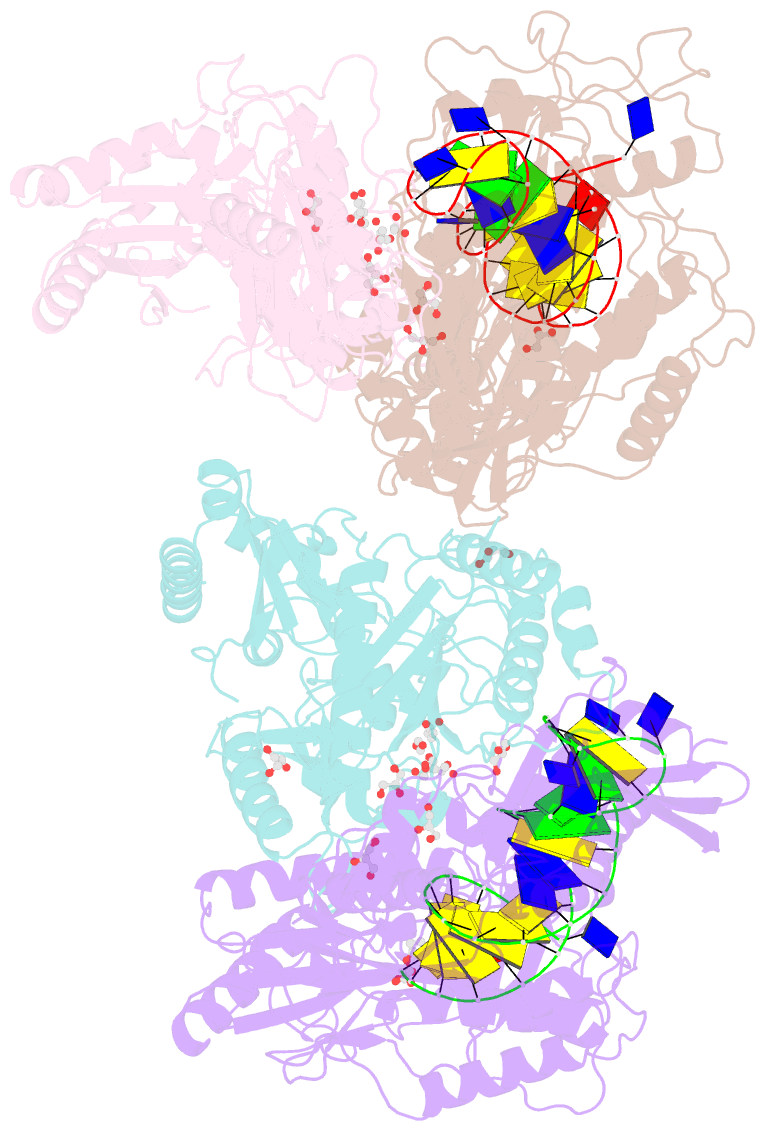
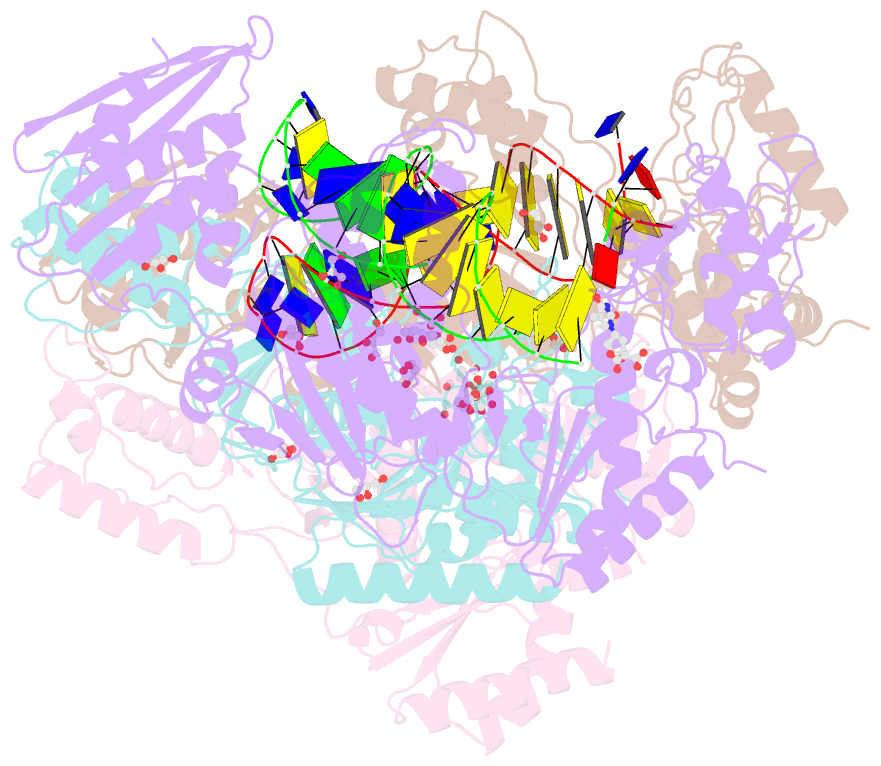
- Structure of hiv-1 reverse transcriptase in complex with a 38-mer hairpin template-primer DNA aptamer and an alpha-carboxyphosphonate inhibitor
- Reference
- Mullins ND, Maguire NM, Ford A, Das K, Arnold E, Balzarini J, Maguire AR (2016): "Exploring the role of the alpha-carboxyphosphonate moiety in the HIV-RT activity of alpha-carboxy nucleoside phosphonates." Org.Biomol.Chem., 14, 2454-2465. doi: 10.1039/c5ob02507a.
- Abstract
- As α-carboxy nucleoside phosphonates (α-CNPs) have demonstrated a novel mode of action of HIV-1 reverse transcriptase inhibition, structurally related derivatives were synthesized, namely the malonate 2, the unsaturated and saturated bisphosphonates 3 and 4, respectively and the amide 5. These compounds were evaluated for inhibition of HIV-1 reverse transcriptase in cell-free assays. The importance of the α-carboxy phosphonoacetic acid moiety for achieving reverse transcriptase inhibition, without the need for prior phosphorylation, was confirmed. The malonate derivative 2 was less active by two orders of magnitude than the original α-CNPs, while displaying the same pattern of kinetic behavior; interestingly the activity resides in the “L”-enantiomer of 2, as seen with the earlier series of α-CNPs. A crystal structure with an RT/DNA complex at 2.95 Å resolution revealed the binding of the “L”-enantiomer of 2, at the polymerase active site with a weaker metal ion chelation environment compared to 1a (T-α-CNP) which may explain the lower inhibitory activity of 2.