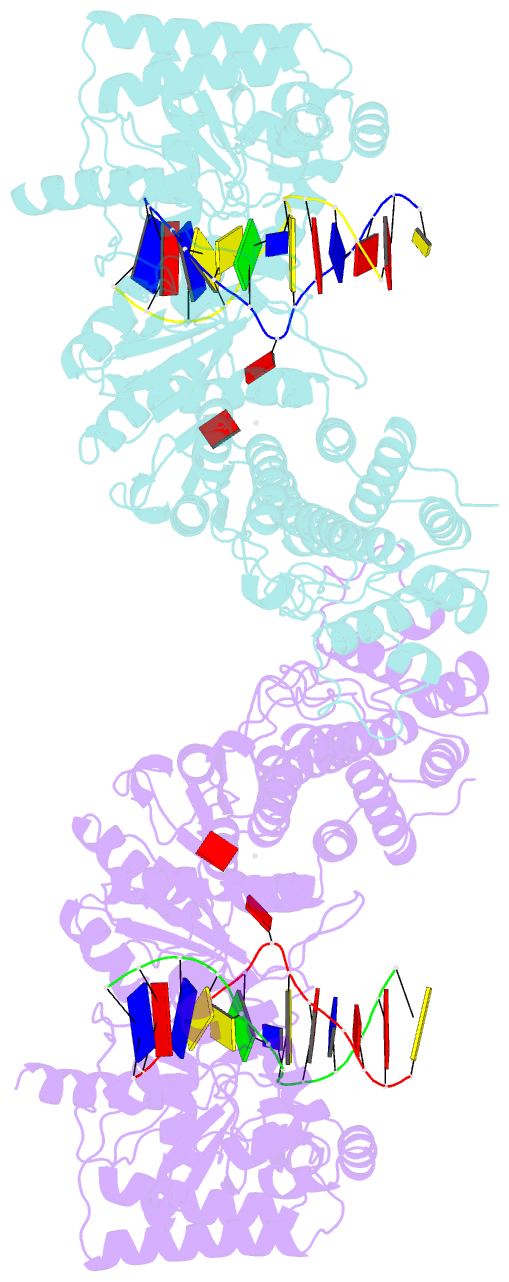
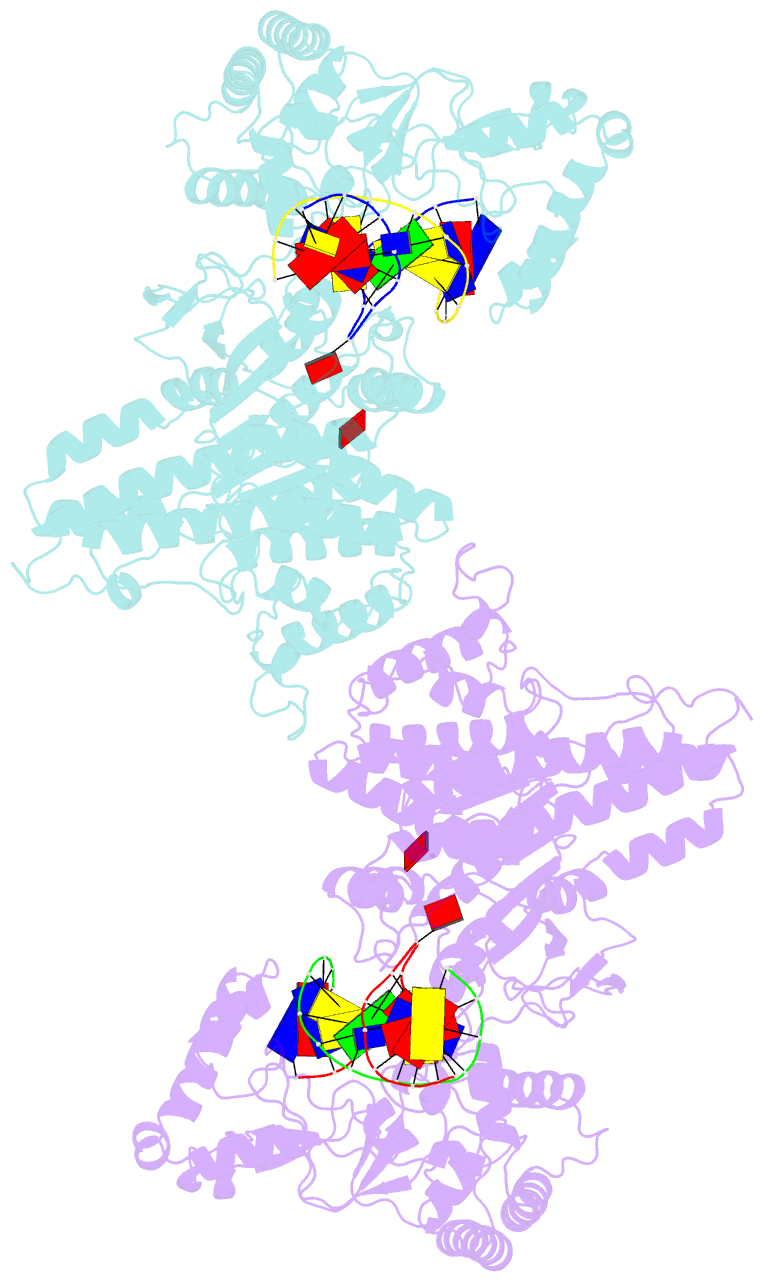
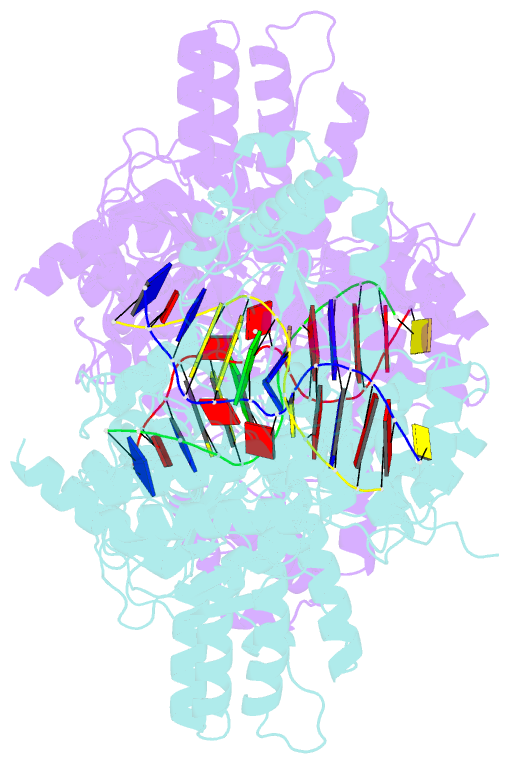
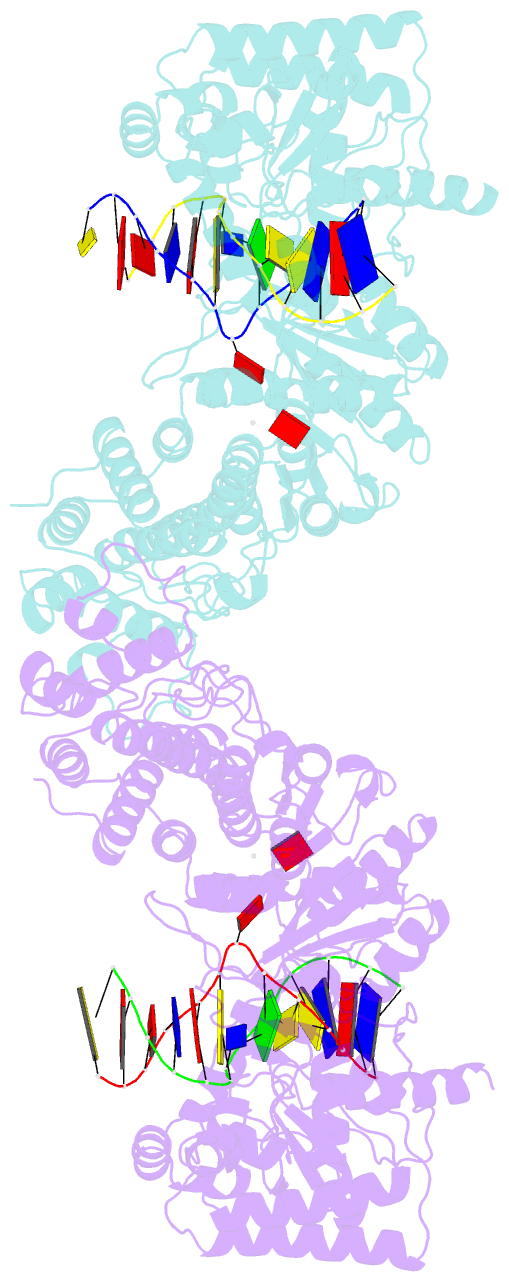
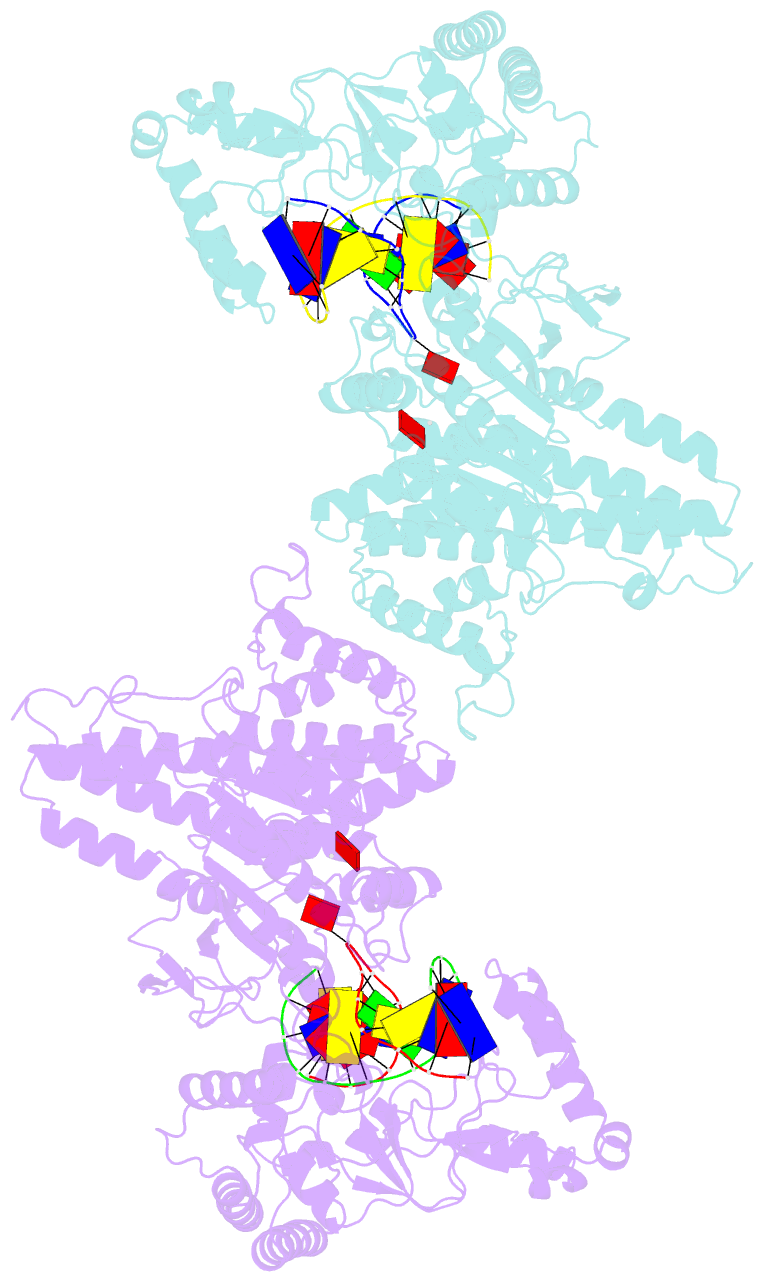
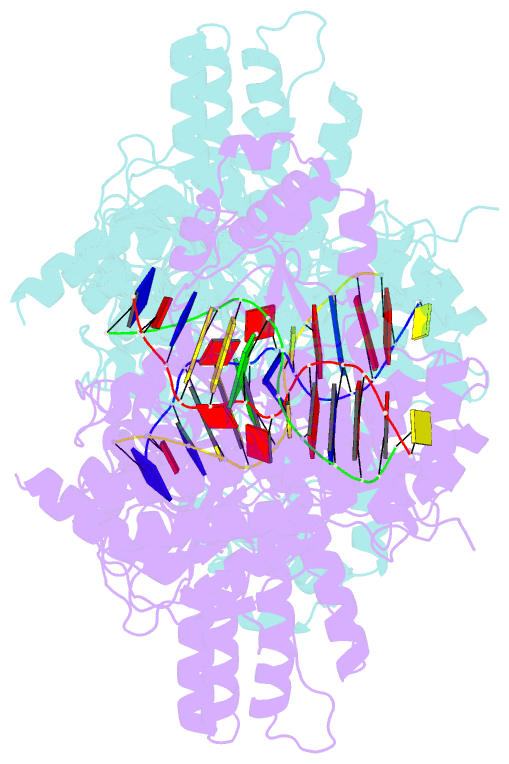
Summary information and primary citation
- PDB-id
- 5hr4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.6 Å)
- Summary
- Structure of type iil restriction-modification enzyme mmei in complex with DNA has implications for engineering of new specificities
- Reference
- Callahan SJ, Luyten YA, Gupta YK, Wilson GG, Roberts RJ, Morgan RD, Aggarwal AK (2016): "Structure of Type IIL Restriction-Modification Enzyme MmeI in Complex with DNA Has Implications for Engineering New Specificities." Plos Biol., 14, e1002442. doi: 10.1371/journal.pbio.1002442.
- Abstract
- The creation of restriction enzymes with programmable DNA-binding and -cleavage specificities has long been a goal of modern biology. The recently discovered Type IIL MmeI family of restriction-and-modification (RM) enzymes that possess a shared target recognition domain provides a framework for engineering such new specificities. However, a lack of structural information on Type IIL enzymes has limited the repertoire that can be rationally engineered. We report here a crystal structure of MmeI in complex with its DNA substrate and an S-adenosylmethionine analog (Sinefungin). The structure uncovers for the first time the interactions that underlie MmeI-DNA recognition and methylation (5'-TCCRAC-3'; R = purine) and provides a molecular basis for changing specificity at four of the six base pairs of the recognition sequence (5'-TCCRAC-3'). Surprisingly, the enzyme is resilient to specificity changes at the first position of the recognition sequence (5'-TCCRAC-3'). Collectively, the structure provides a basis for engineering further derivatives of MmeI and delineates which base pairs of the recognition sequence are more amenable to alterations than others.