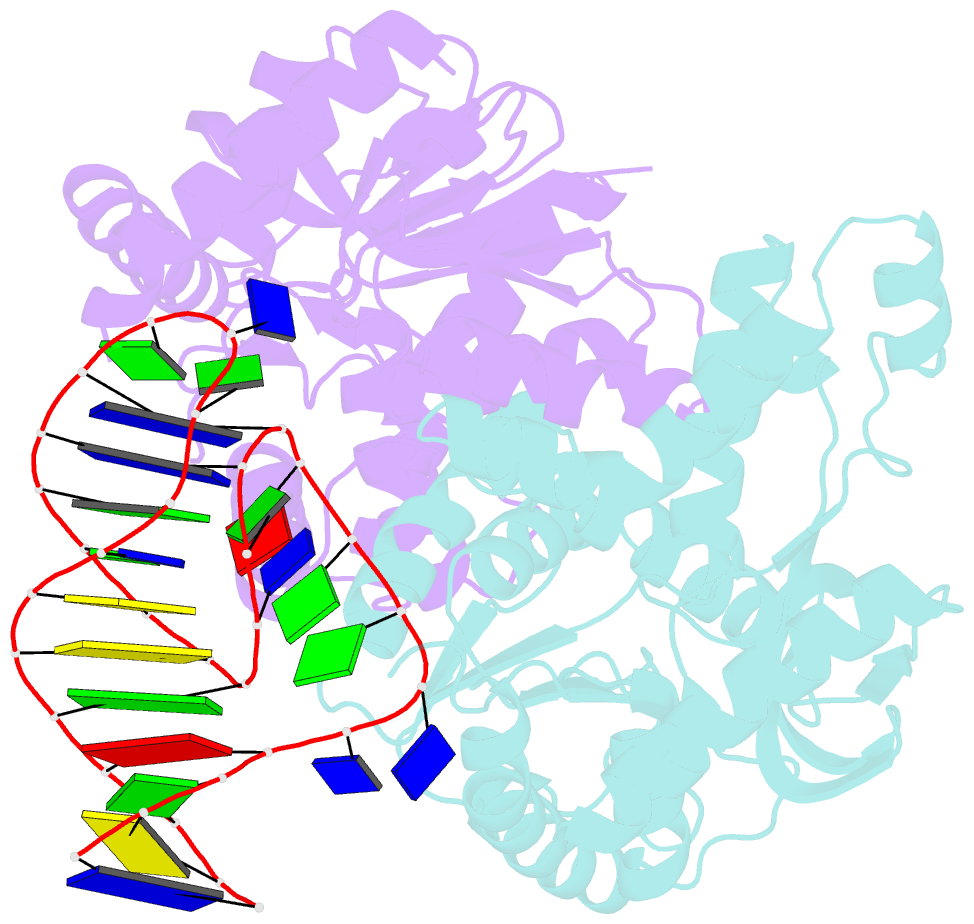
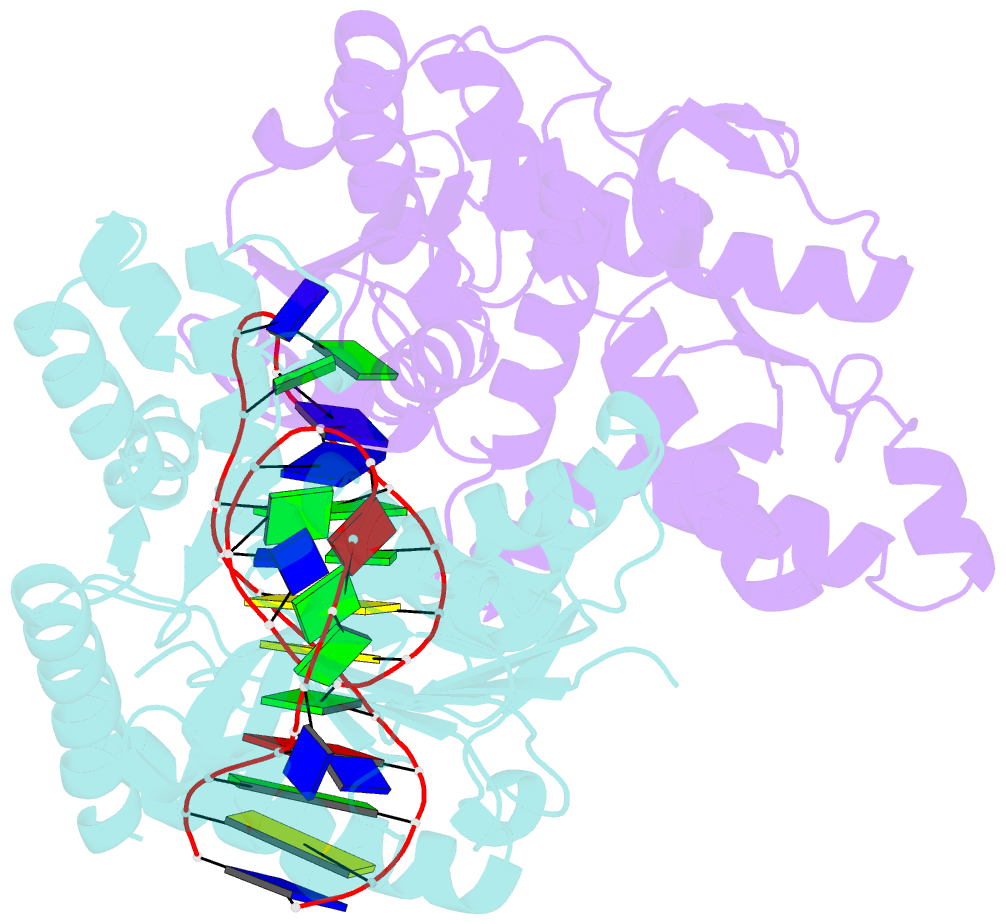
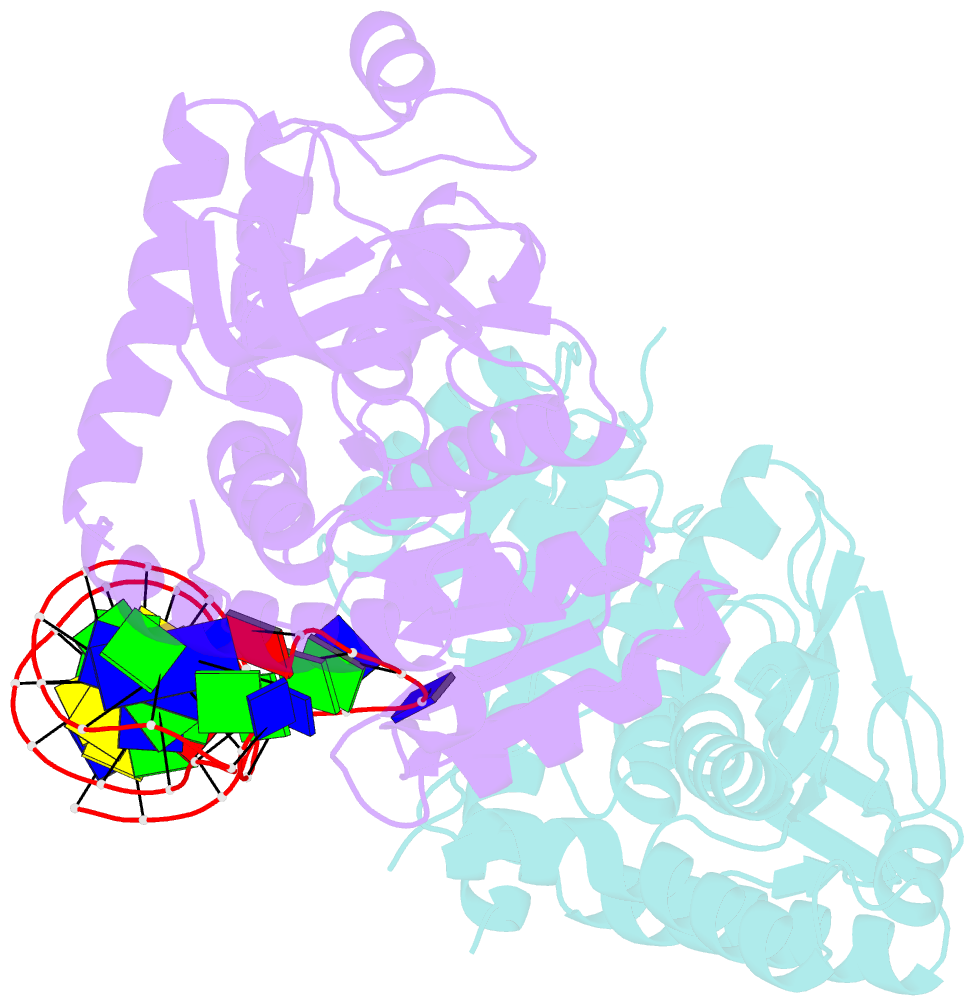
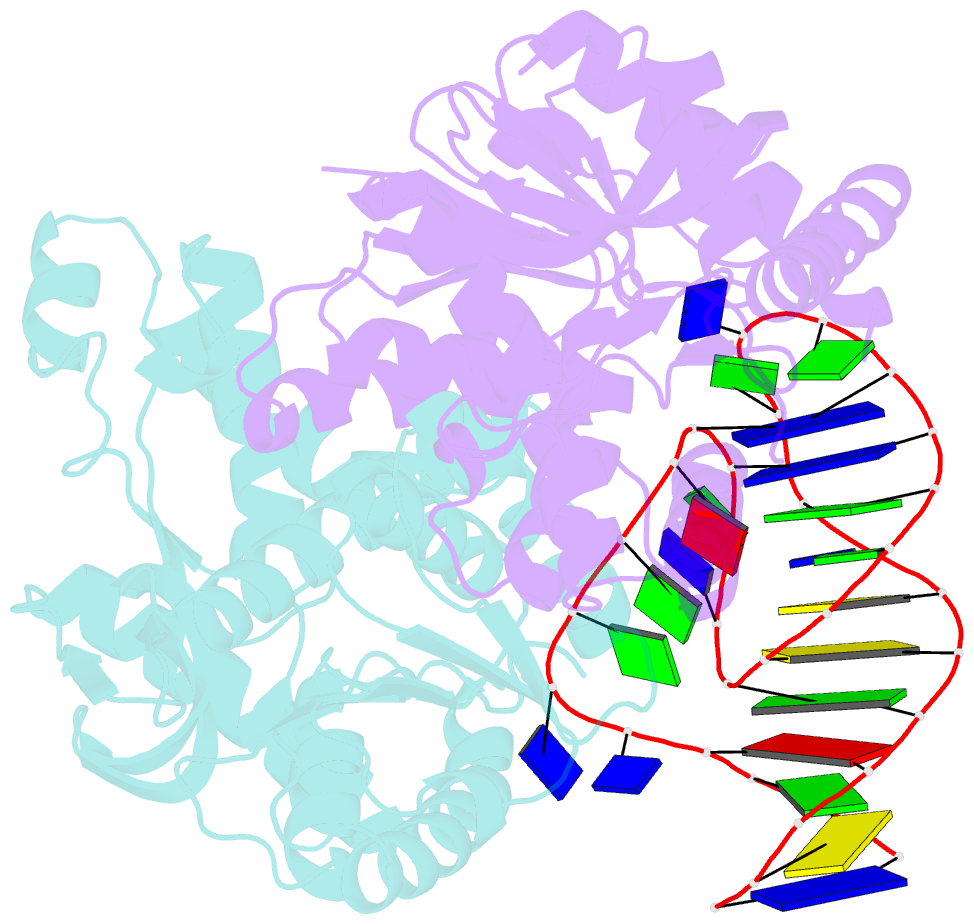
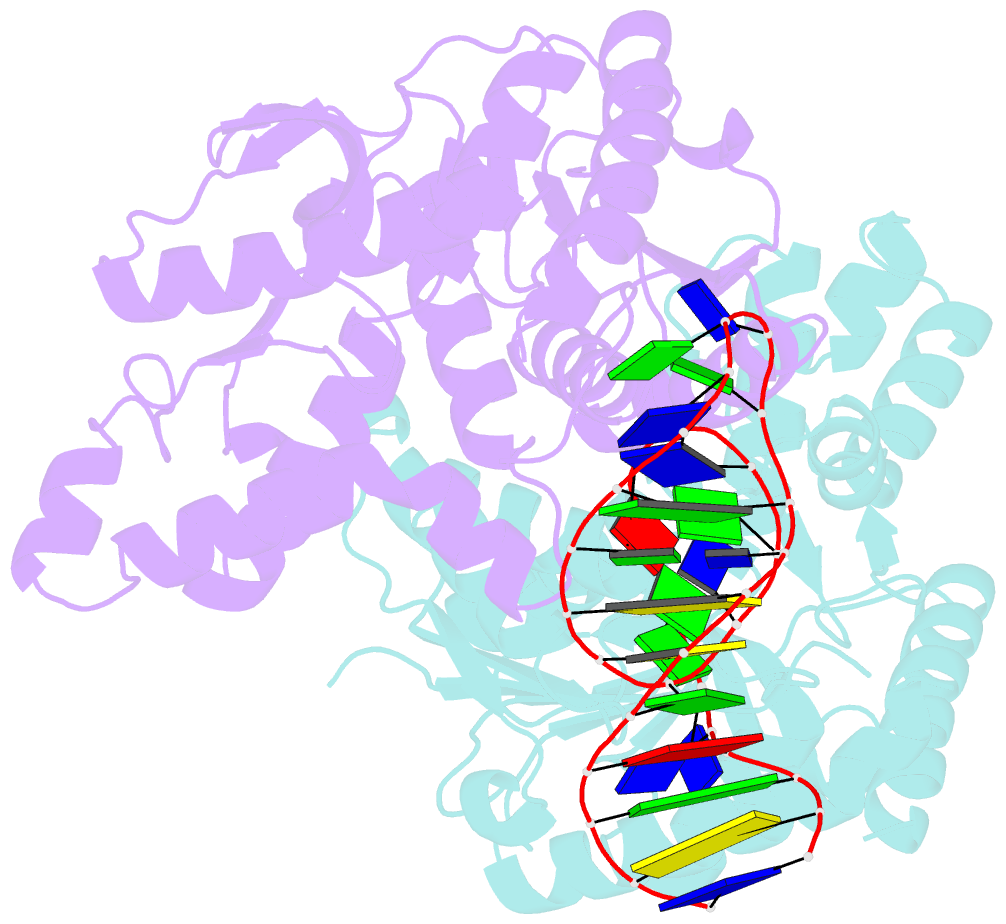
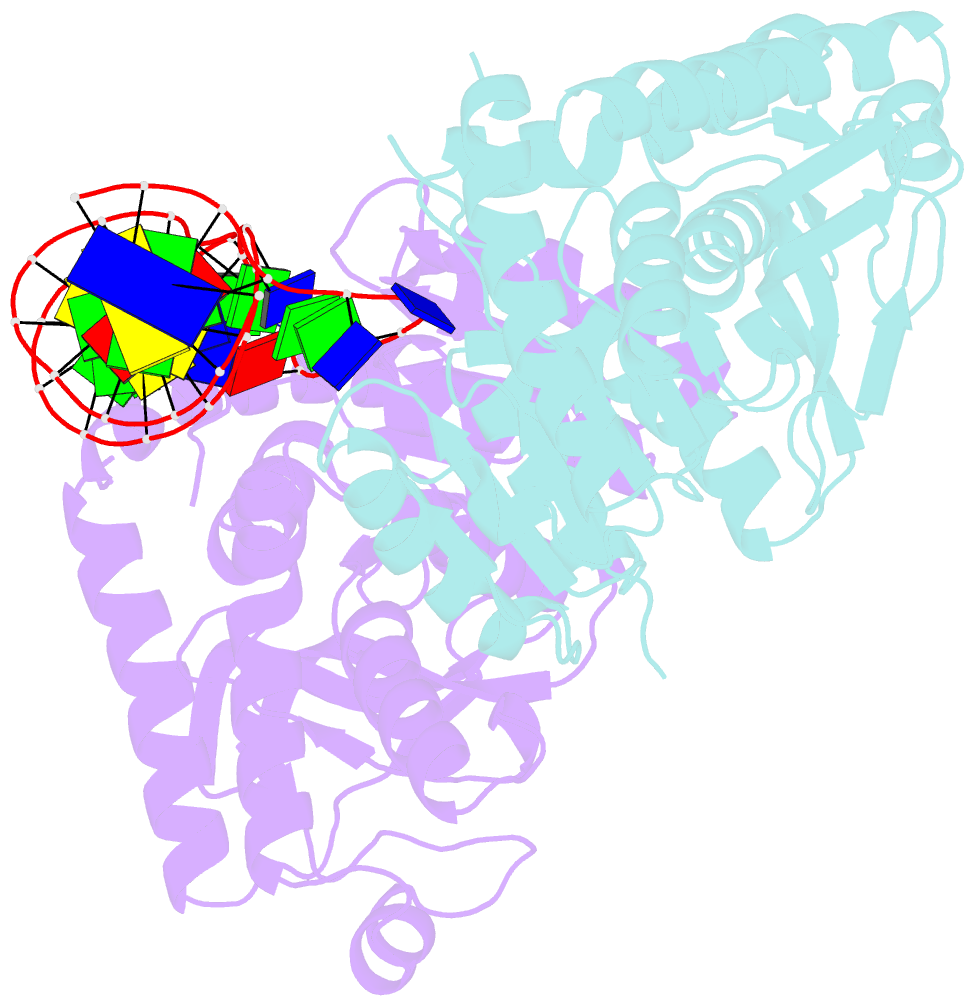
Summary information and primary citation
- PDB-id
- 5hru; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- oxidoreductase-DNA
- Method
- X-ray (1.71 Å)
- Summary
- Crystal structure of plasmodium vivax ldh in complex with a DNA aptamer called pl1
- Reference
- Choi SJ, Ban C (2016): "Crystal structure of a DNA aptamer bound to PvLDH elucidates novel single-stranded DNA structural elements for folding and recognition." Sci Rep, 6, 34998. doi: 10.1038/srep34998.
- Abstract
- Structural elements are key elements for understanding single-stranded nucleic acid folding. Although various RNA structural elements have been documented, structural elements of single-stranded DNA (ssDNA) have rarely been reported. Herein, we determined a crystal structure of PvLDH in complex with a DNA aptamer called pL1. This aptamer folds into a hairpin-bulge contact by adopting three novel structural elements, viz, DNA T-loop-like motif, base-phosphate zipper, and DNA G·G metal ion zipper. Moreover, the pL1:PvLDH complex shows unique properties compared with other protein:nucleic acid complexes. Generally, extensive intermolecular hydrogen bonds occur between unpaired nucleotides and proteins for specific recognitions. Although most protein-interacting nucleotides of pL1 are unpaired nucleotides, pL1 recognizes PvLDH by predominant shape complementarity with many bridging water molecules owing to the combination of three novel structural elements making protein-binding unpaired nucleotides stable. Moreover, the additional set of Plasmodium LDH residues which were shown to form extensive hydrogen bonds with unpaired nucleotides of 2008s does not participate in the recognition of pL1. Superimposition of the pL1:PvLDH complex with hLDH reveals steric clashes between pL1 and hLDH in contrast with no steric clashes between 2008s and hLDH. Therefore, specific protein recognition mode of pL1 is totally different from that of 2008s.