Summary information and primary citation
- PDB-id
- 5i2d; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA-RNA
- Method
- X-ray (4.405 Å)
- Summary
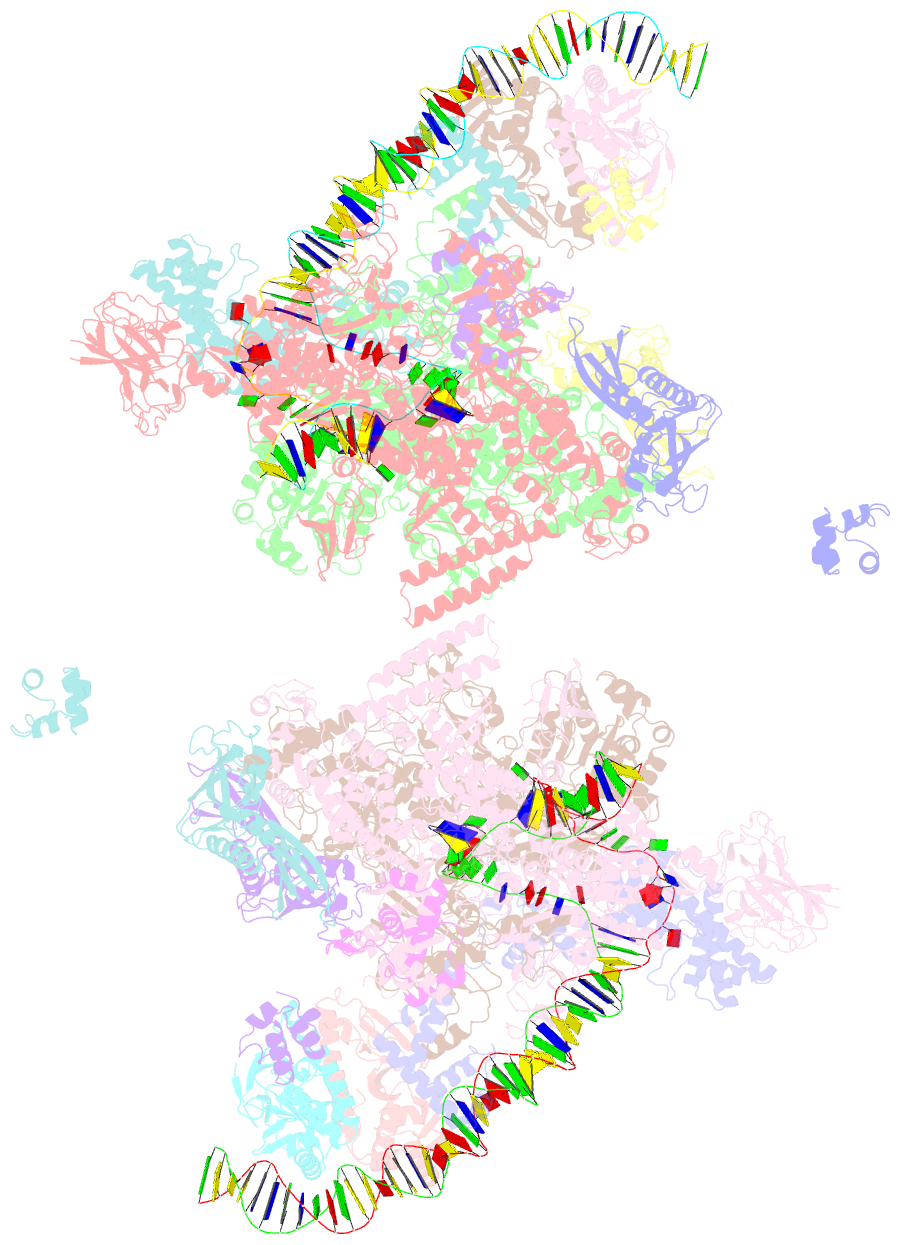
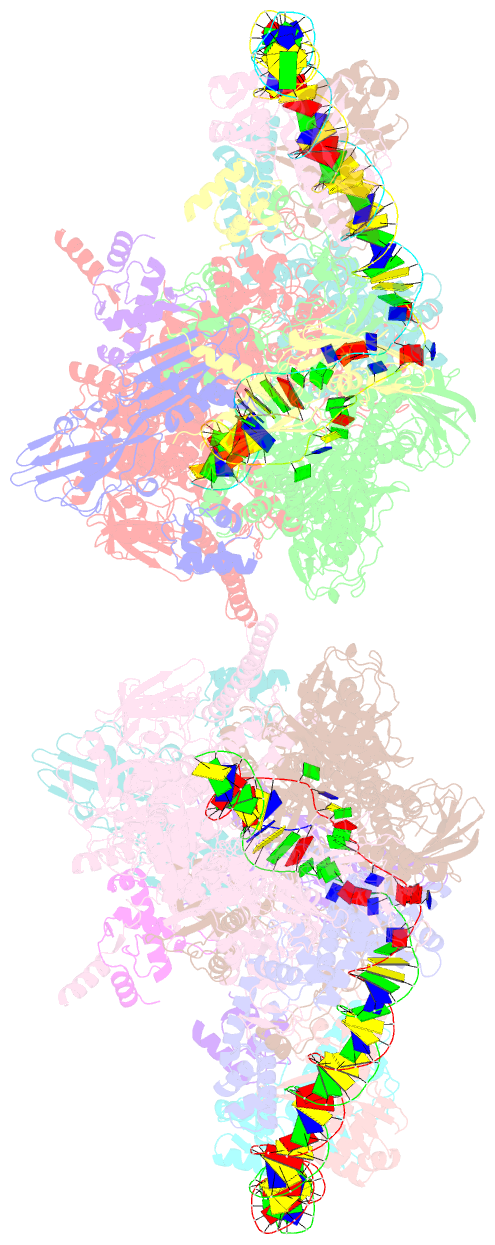
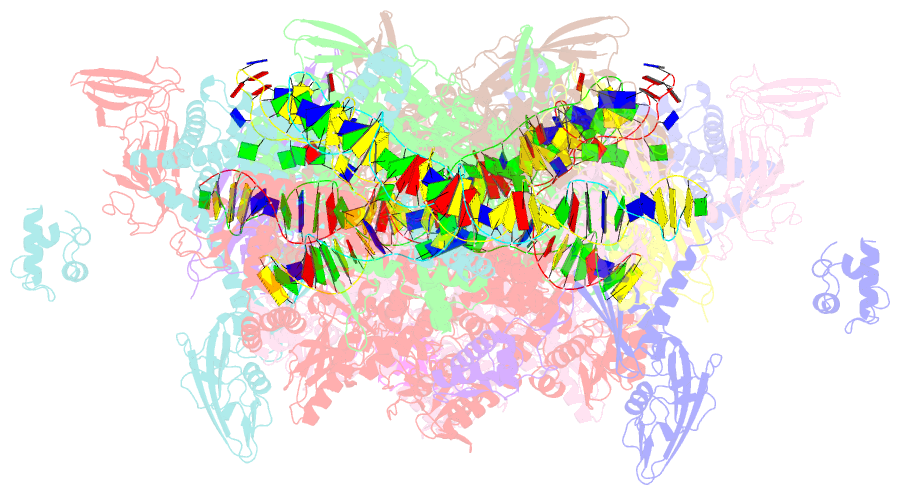
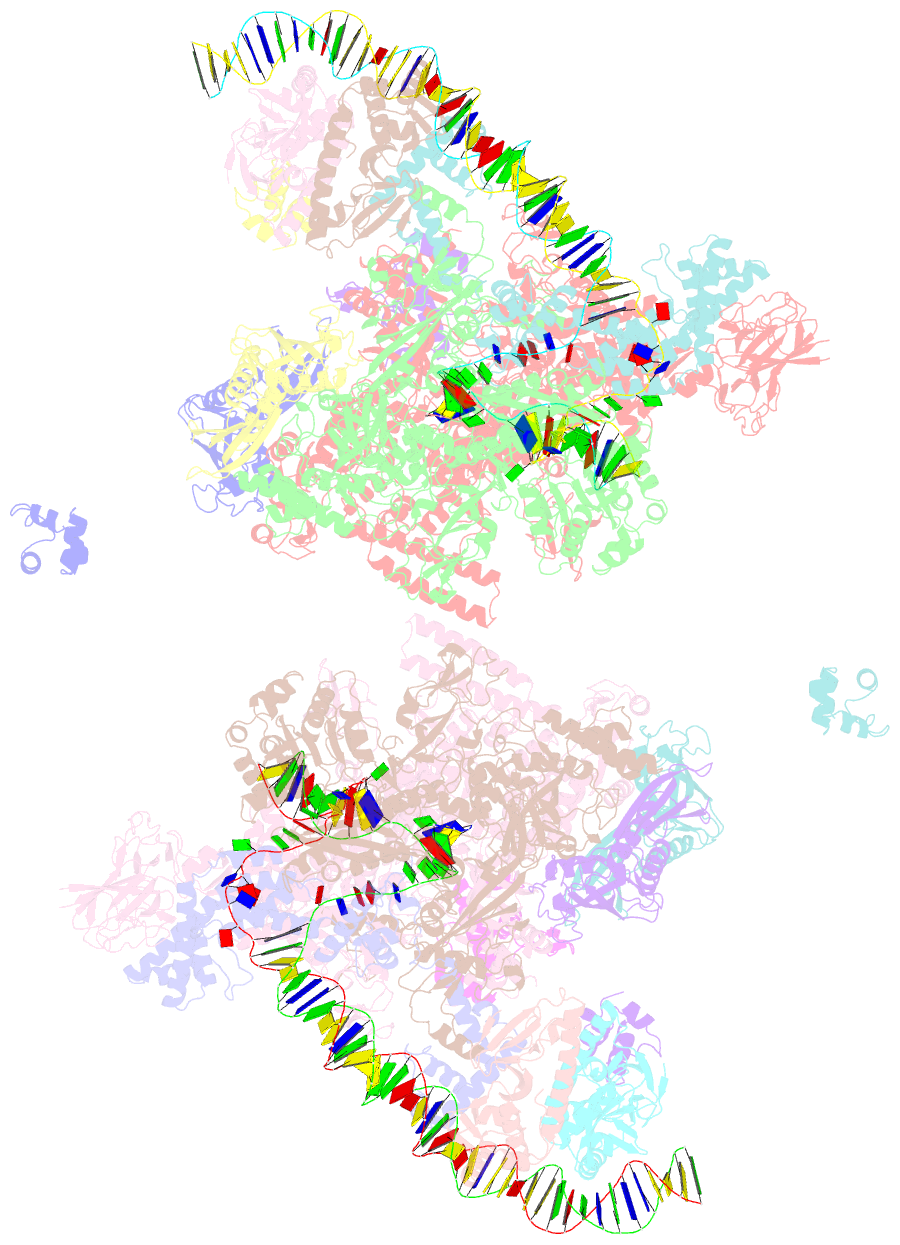
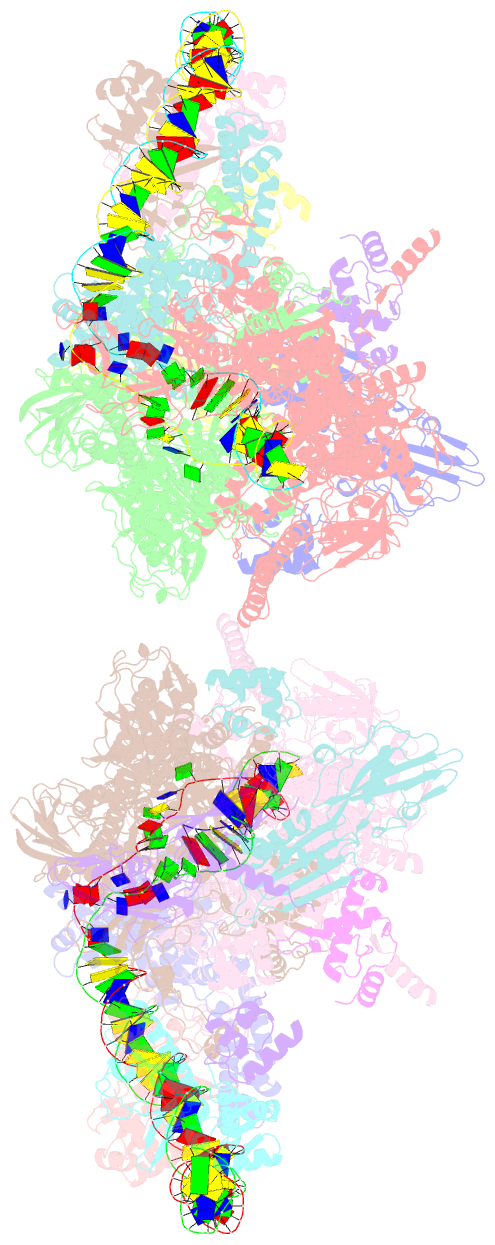
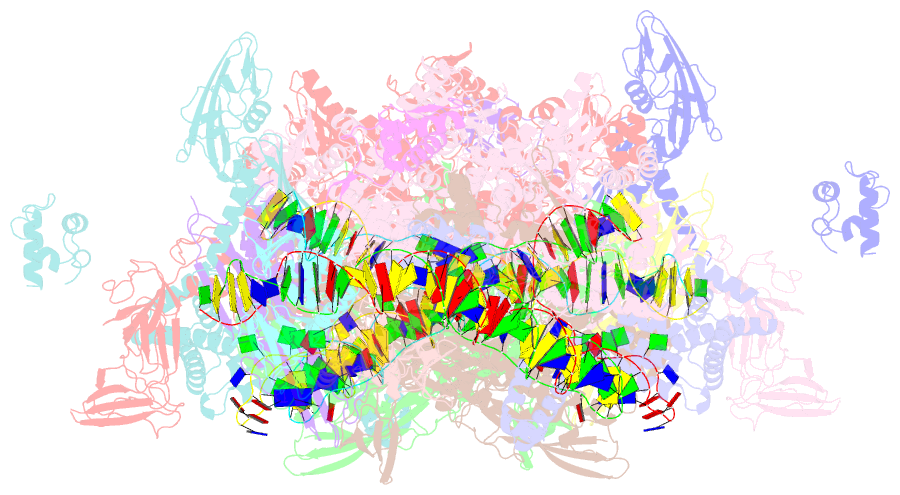
- Crystal structure of t. thermophilus tthb099 class ii transcription activation complex: tap-rpo
- Reference
- Feng Y, Zhang Y, Ebright RH (2016): "Structural basis of transcription activation." Science, 352, 1330-1333. doi: 10.1126/science.aaf4417.
- Abstract
- Class II transcription activators function by binding to a DNA site overlapping a core promoter and stimulating isomerization of an initial RNA polymerase (RNAP)-promoter closed complex into a catalytically competent RNAP-promoter open complex. Here, we report a 4.4 angstrom crystal structure of an intact bacterial class II transcription activation complex. The structure comprises Thermus thermophilus transcription activator protein TTHB099 (TAP) [homolog of Escherichia coli catabolite activator protein (CAP)], T. thermophilus RNAP σ(A) holoenzyme, a class II TAP-dependent promoter, and a ribotetranucleotide primer. The structure reveals the interactions between RNAP holoenzyme and DNA responsible for transcription initiation and reveals the interactions between TAP and RNAP holoenzyme responsible for transcription activation. The structure indicates that TAP stimulates isomerization through simple, adhesive, stabilizing protein-protein interactions with RNAP holoenzyme.