Summary information and primary citation
- PDB-id
- 5i44; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.621 Å)
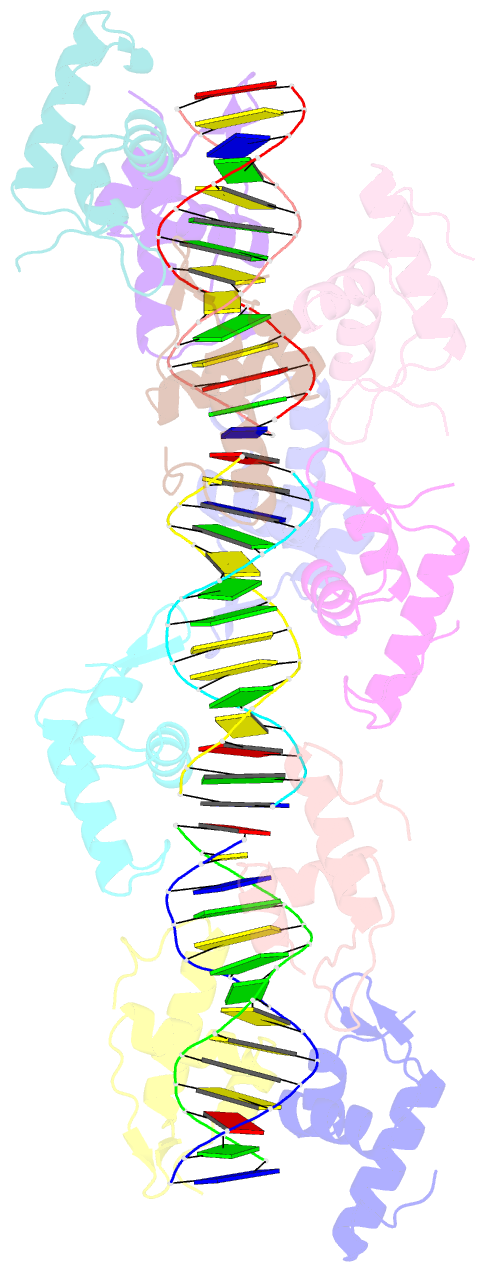
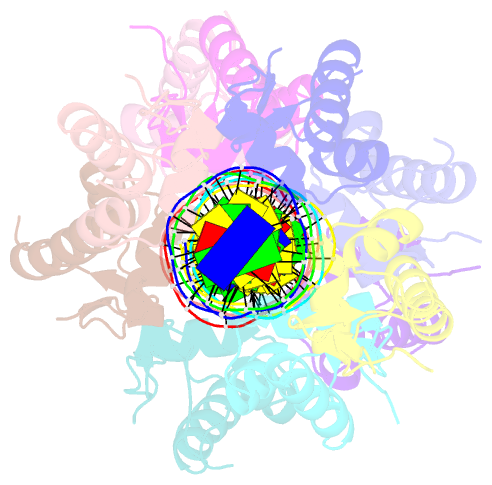
- Summary
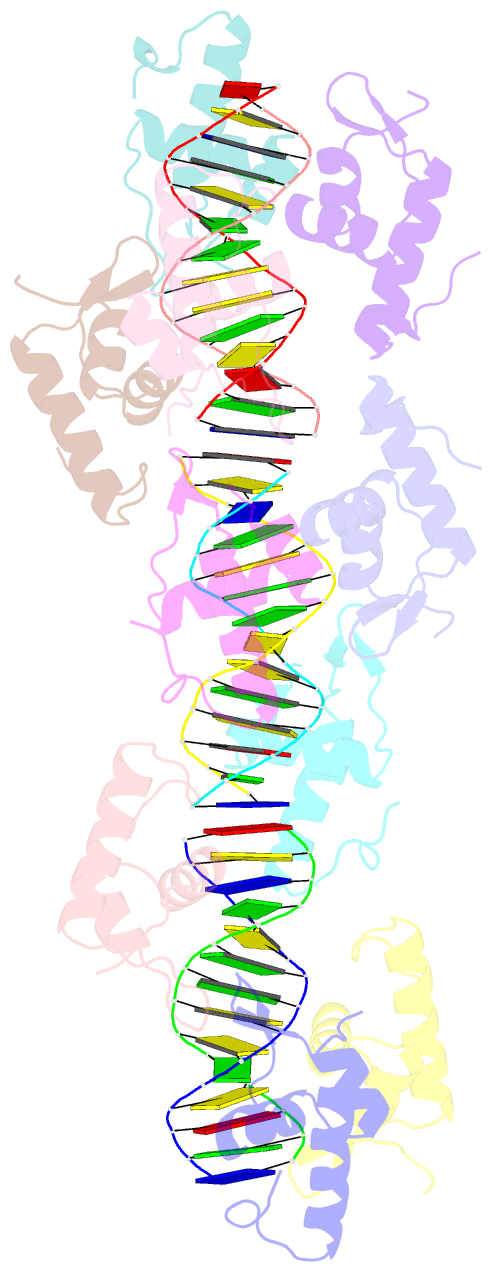
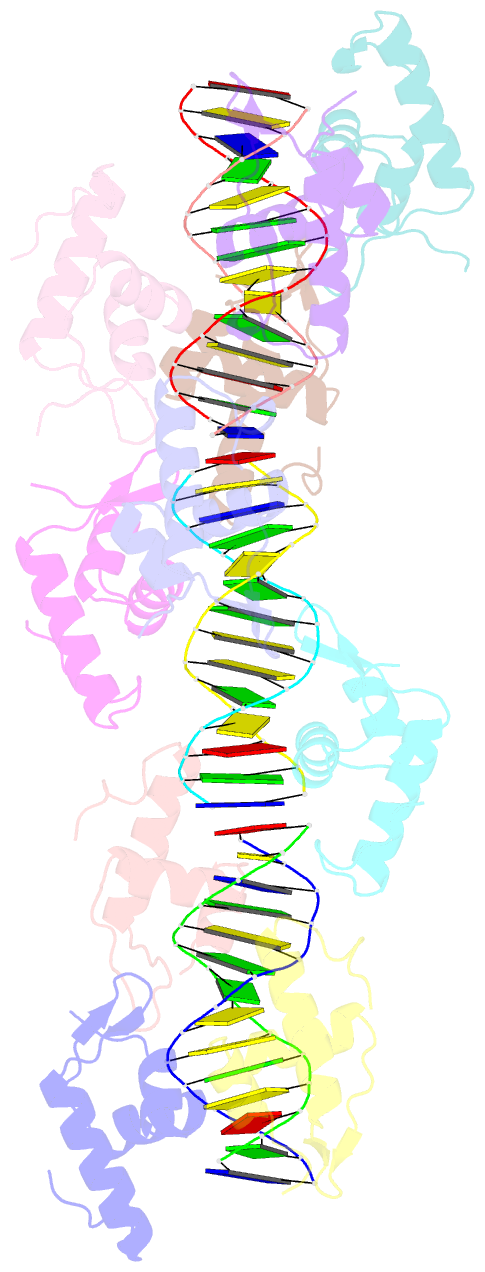
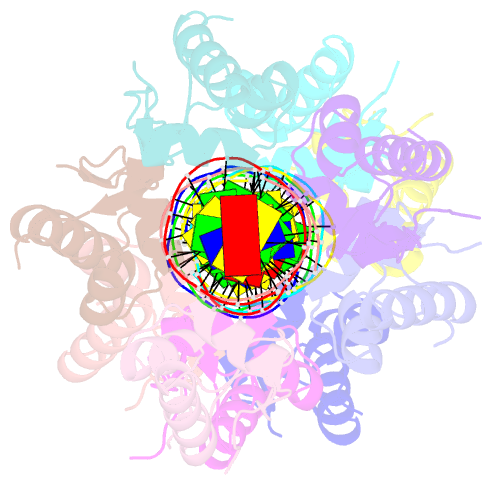
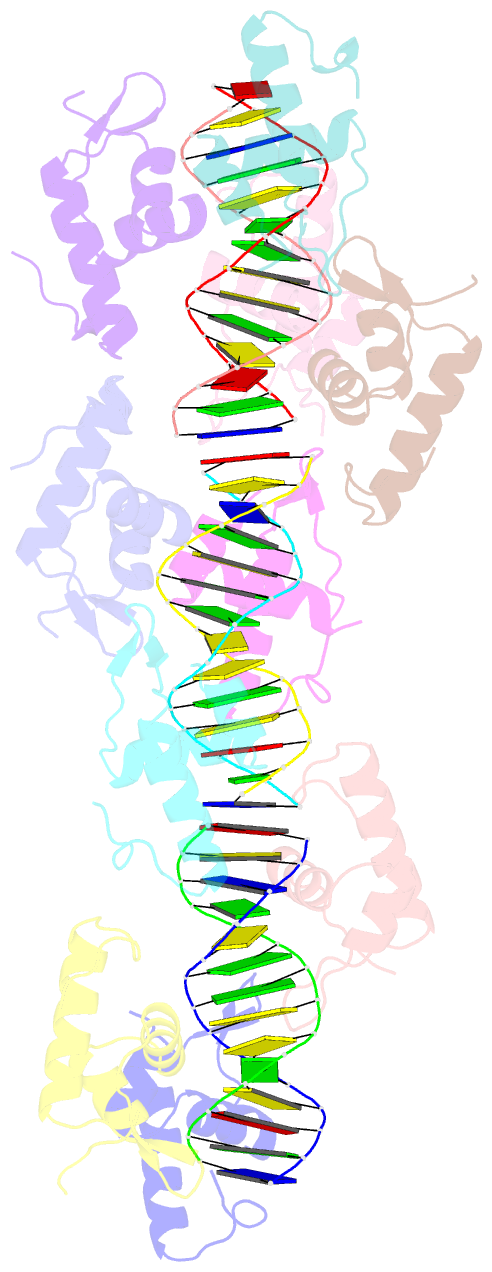
- Structure of raca-DNA complex; p21 form
- Reference
- Schumacher MA, Lee J, Zeng W (2016): "Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein." Nucleic Acids Res., 44, 5438-5449. doi: 10.1093/nar/gkw248.
- Abstract
- During Bacillus subtilis sporulation, segregating sister chromosomes are anchored to cell poles and the chromosome is remodeled into an elongated structure called the axial filament. Data indicate that a developmentally regulated protein called RacA is involved in these functions. To gain insight into how RacA performs these diverse processes we performed a battery of structural and biochemical analyses. These studies show that RacA contains an N-terminal winged-helix-turn-helix module connected by a disordered region to a predicted coiled-coil domain. Structures capture RacA binding the DNA using distinct protein-protein interfaces and employing adjustable DNA docking modes. This unique DNA binding mechanism indicates how RacA can both specifically recognize its GC-rich centromere and also non-specifically bind the DNA. Adjacent RacA molecules within the protein-DNA structure interact leading to DNA compaction, suggesting a mechanism for axial filament formation. We also show that the RacA C-domain coiled coil directly contacts the coiled coil region of the polar protein DivIVA, which anchors RacA and hence the chromosome to the pole. Thus, our combined data reveal unique DNA binding properties by RacA and provide insight into the DNA remodeling and polar anchorage functions of the protein.