Summary information and primary citation
- PDB-id
- 5i50; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.701 Å)
- Summary
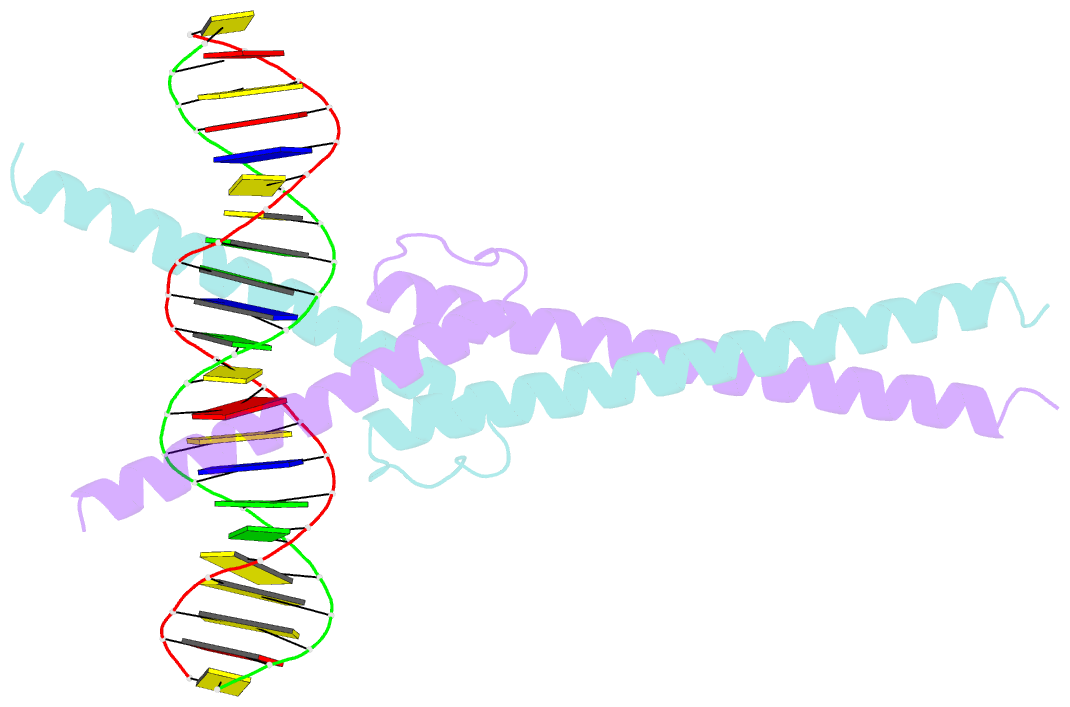
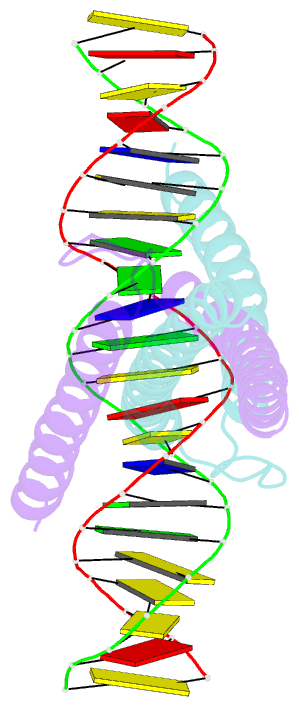
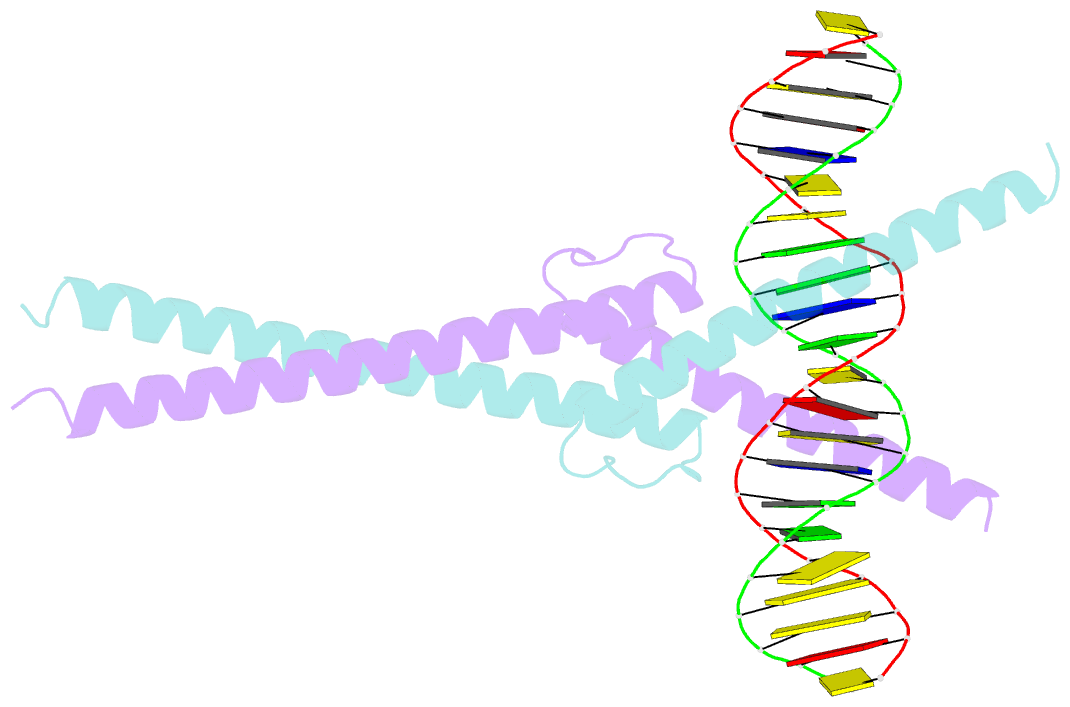
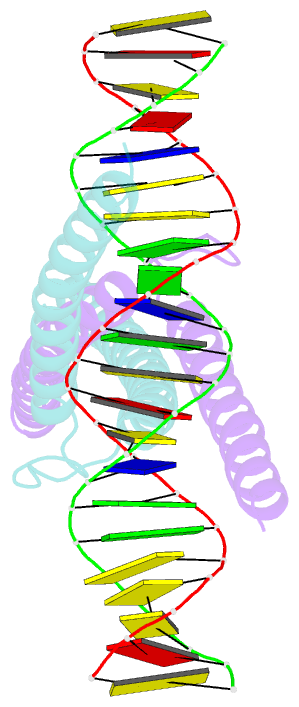
- Structure of omomyc bound to double-stranded DNA
- Reference
- Jung LA, Gebhardt A, Koelmel W, Ade CP, Walz S, Kuper J, von Eyss B, Letschert S, Redel C, d'Artista L, Biankin A, Zender L, Sauer M, Wolf E, Evan G, Kisker C, Eilers M (2017): "OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors." Oncogene, 36, 1911-1924. doi: 10.1038/onc.2016.354.
- Abstract
- MYC genes have both essential roles during normal development and exert oncogenic functions during tumorigenesis. Expression of a dominant-negative allele of MYC, termed OmoMYC, can induce rapid tumor regression in mouse models with little toxicity for normal tissues. How OmoMYC discriminates between physiological and oncogenic functions of MYC is unclear. We have solved the crystal structure of OmoMYC and show that it forms a stable homodimer and as such recognizes DNA in the same manner as the MYC/MAX heterodimer. OmoMYC attenuates both MYC-dependent activation and repression by competing with MYC/MAX for binding to chromatin, effectively lowering MYC/MAX occupancy at its cognate binding sites. OmoMYC causes the largest decreases in promoter occupancy and changes in expression on genes that are invaded by oncogenic MYC levels. A signature of OmoMYC-regulated genes defines subgroups with high MYC levels in multiple tumor entities and identifies novel targets for the eradication of MYC-driven tumors.