Summary information and primary citation
- PDB-id
- 5i9d; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.596 Å)
- Summary
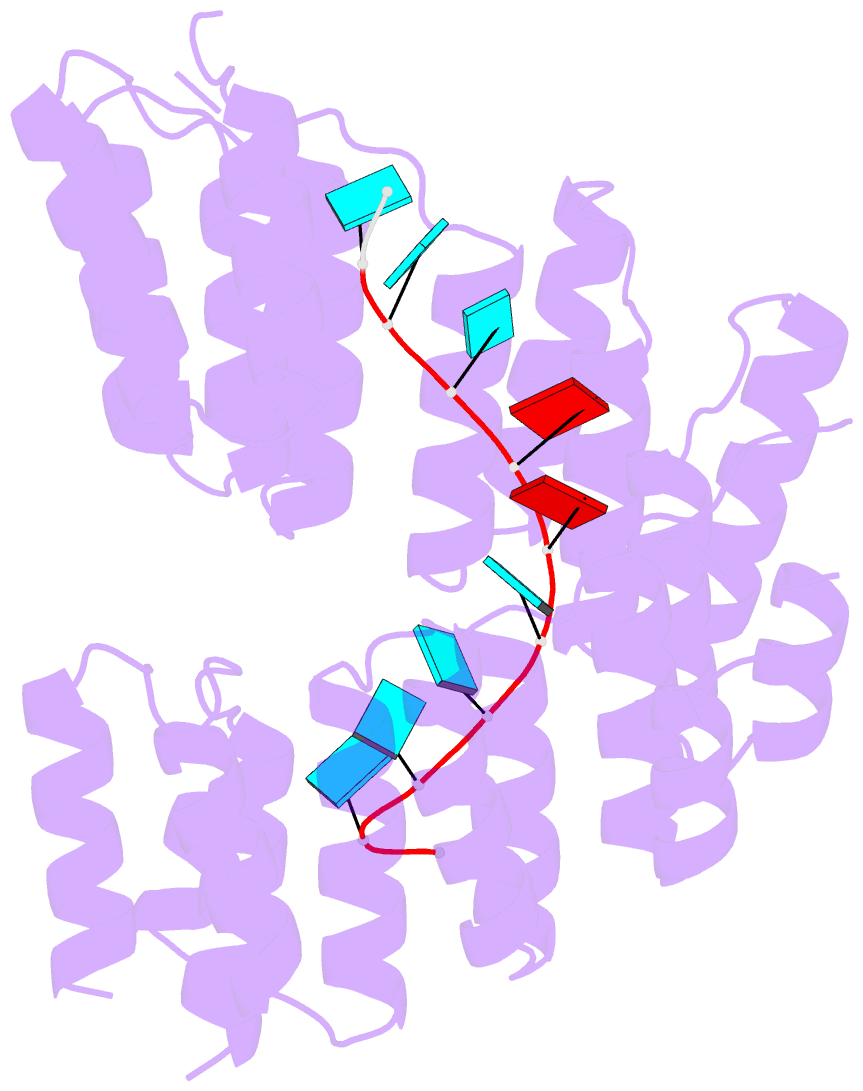
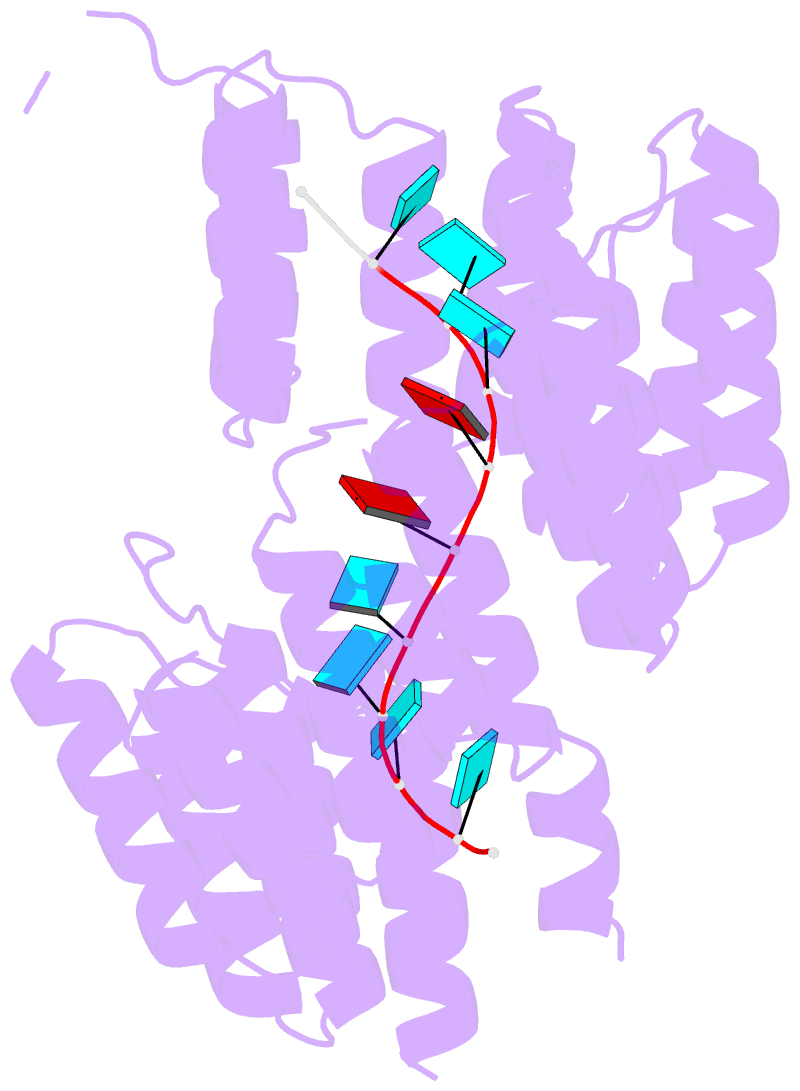
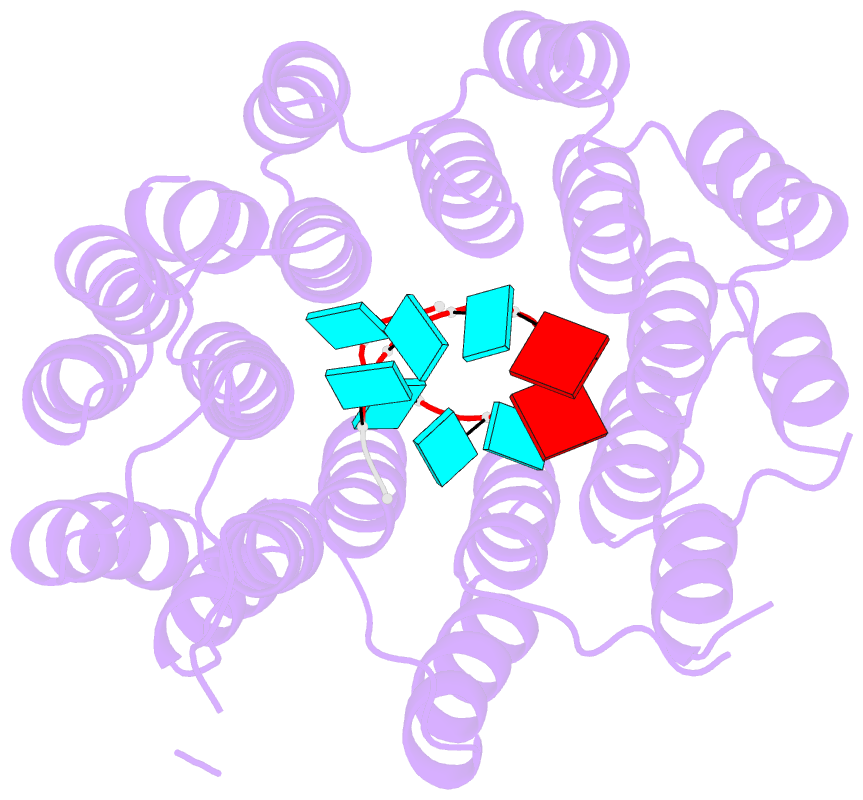
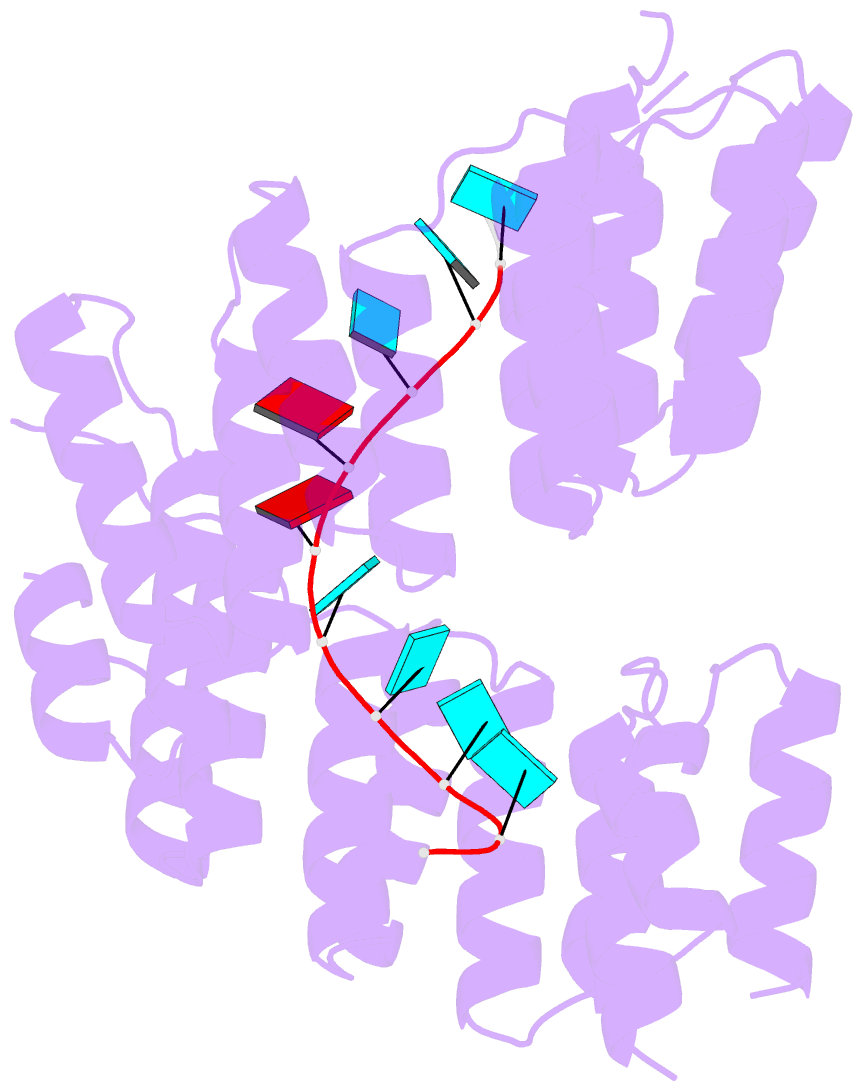
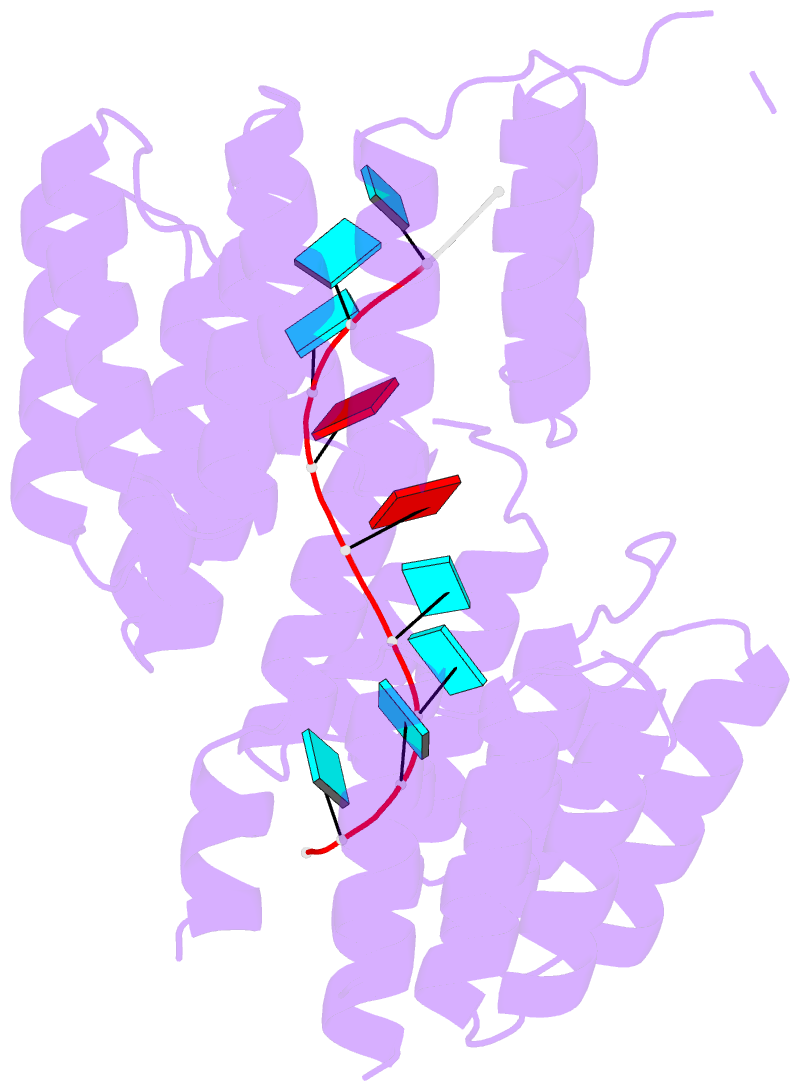
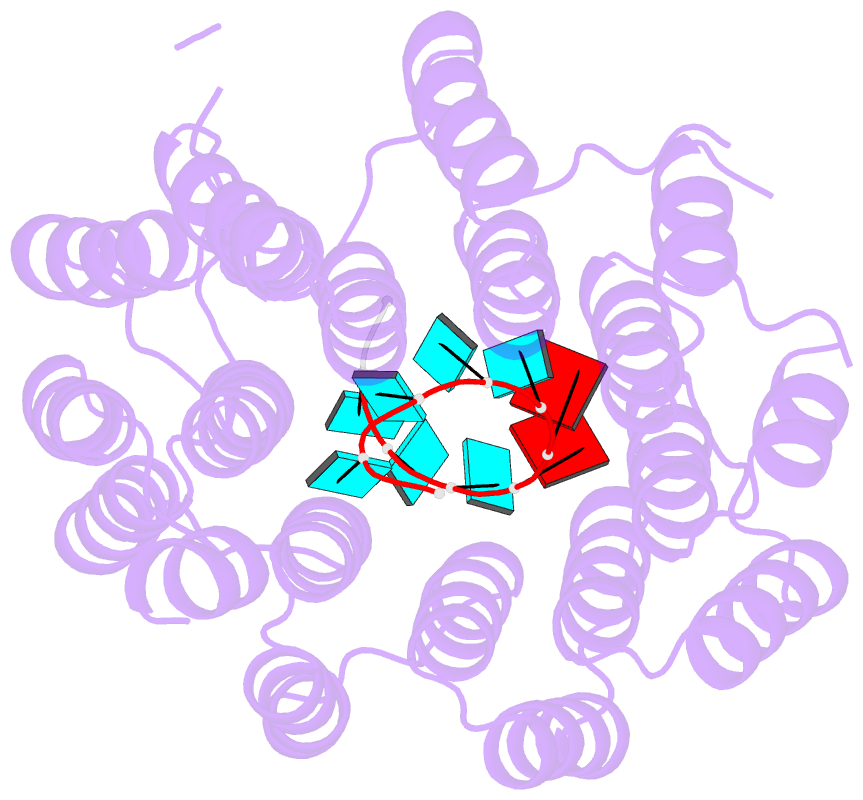
- Crystal structure of designed pentatricopeptide repeat protein dppr-u8a2 in complex with its target RNA u8a2
- Reference
- Shen C, Zhang D, Guan Z, Liu Y, Yang Z, Yang Y, Wang X, Wang Q, Zhang Q, Fan S, Zou T, Yin P (2016): "Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins." Nat Commun, 7, 11285. doi: 10.1038/ncomms11285.
- Abstract
- As a large family of RNA-binding proteins, pentatricopeptide repeat (PPR) proteins mediate multiple aspects of RNA metabolism in eukaryotes. Binding to their target single-stranded RNAs (ssRNAs) in a modular and base-specific fashion, PPR proteins can serve as designable modules for gene manipulation. However, the structural basis for nucleotide-specific recognition by designer PPR (dPPR) proteins remains to be elucidated. Here, we report four crystal structures of dPPR proteins in complex with their respective ssRNA targets. The dPPR repeats are assembled into a right-handed superhelical spiral shell that embraces the ssRNA. Interactions between different PPR codes and RNA bases are observed at the atomic level, revealing the molecular basis for the modular and specific recognition patterns of the RNA bases U, C, A and G. These structures not only provide insights into the functional study of PPR proteins but also open a path towards the potential design of synthetic sequence-specific RNA-binding proteins.