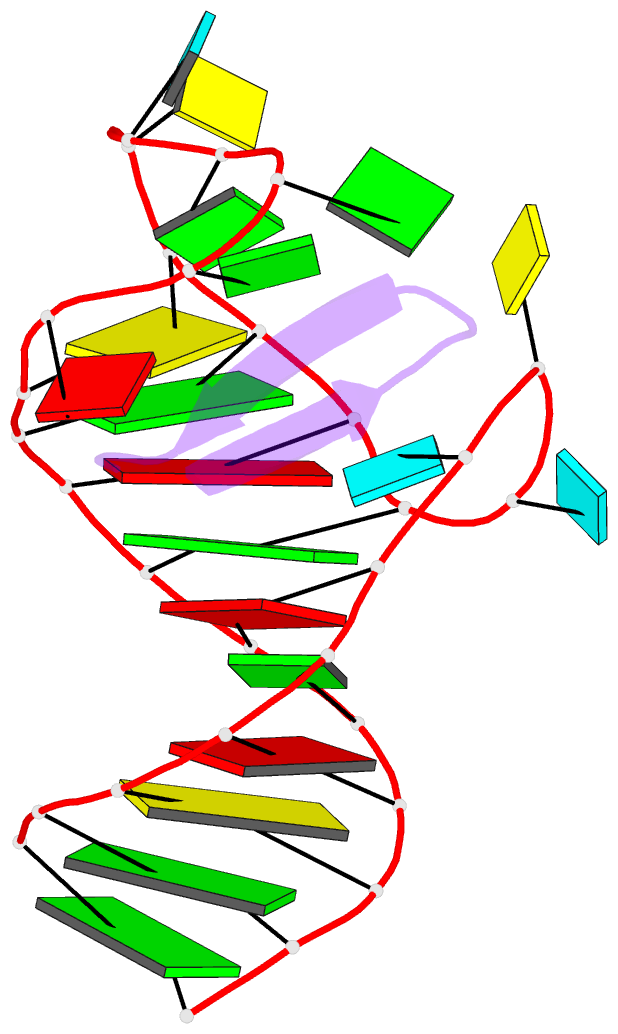
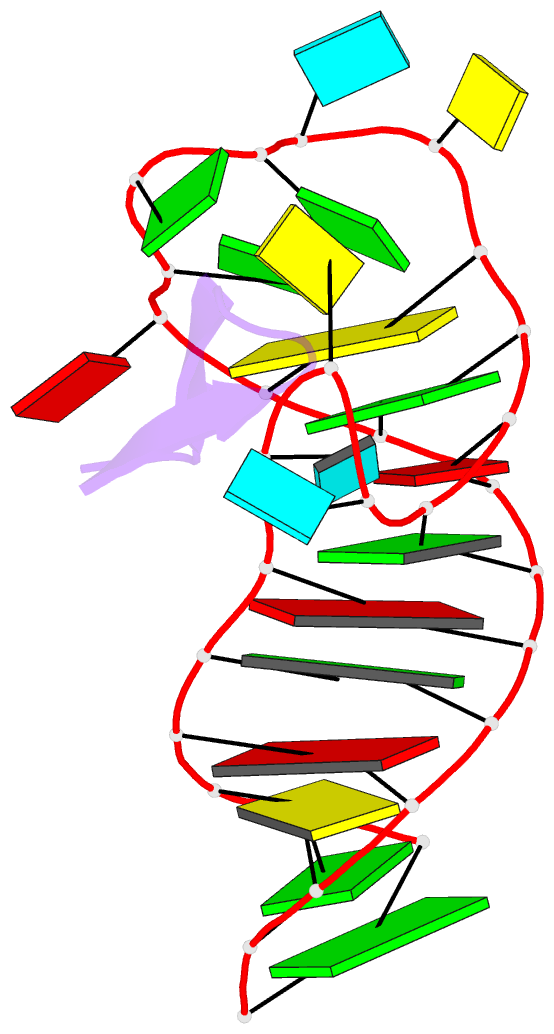
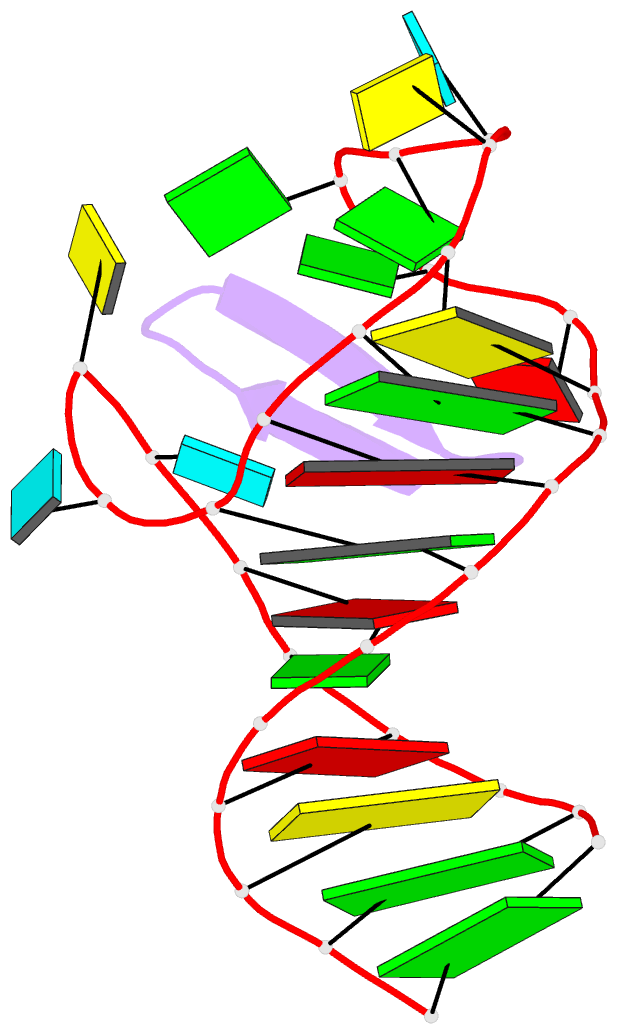
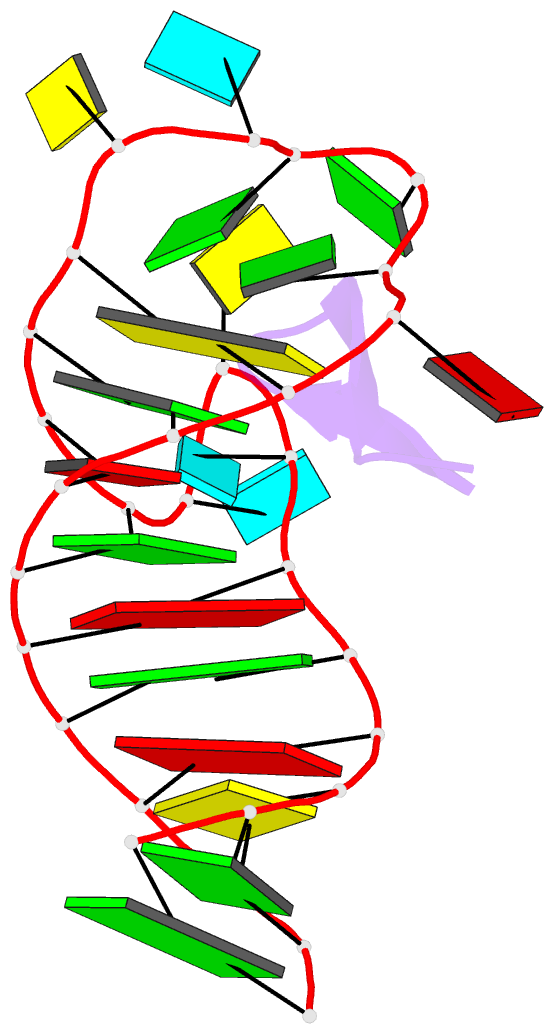
Summary information and primary citation
- PDB-id
- 5j0m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- NMR
- Summary
- Ground state sampled during rdc restrained replica-averaged metadynamics (ram) simulations of the hiv-1 tar complexed with cyclic peptide mimetic of tat
- Reference
- Borkar AN, Bardaro MF, Camilloni C, Aprile FA, Varani G, Vendruscolo M (2016): "Structure of a low-population binding intermediate in protein-RNA recognition." Proc.Natl.Acad.Sci.USA, 113, 7171-7176. doi: 10.1073/pnas.1521349113.
- Abstract
- The interaction of the HIV-1 protein transactivator of transcription (Tat) and its cognate transactivation response element (TAR) RNA transactivates viral transcription and represents a paradigm for the widespread occurrence of conformational rearrangements in protein-RNA recognition. Although the structures of free and bound forms of TAR are well characterized, the conformations of the intermediates in the binding process are still unknown. By determining the free energy landscape of the complex using NMR residual dipolar couplings in replica-averaged metadynamics simulations, we observe two low-population intermediates. We then rationally design two mutants, one in the protein and another in the RNA, that weaken specific nonnative interactions that stabilize one of the intermediates. By using surface plasmon resonance, we show that these mutations lower the release rate of Tat, as predicted. These results identify the structure of an intermediate for RNA-protein binding and illustrate a general strategy to achieve this goal with high resolution.