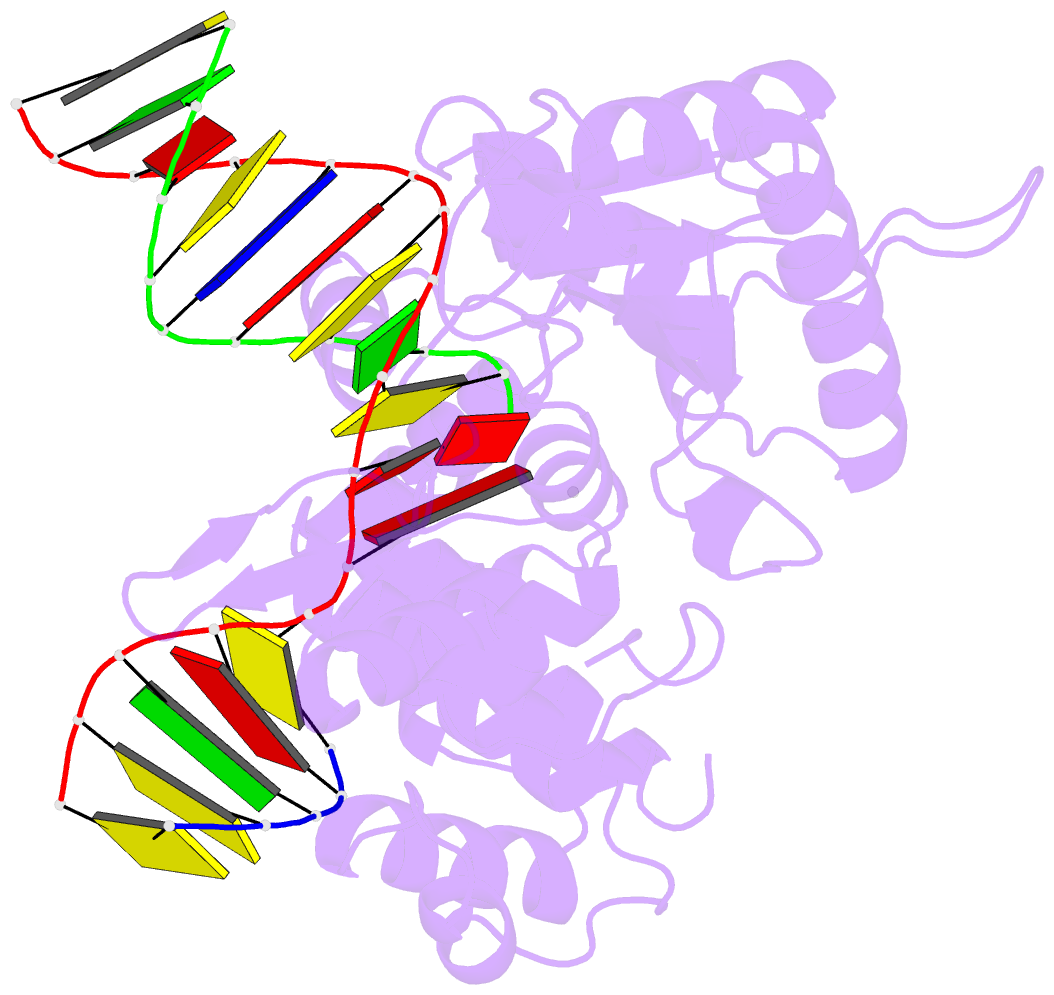
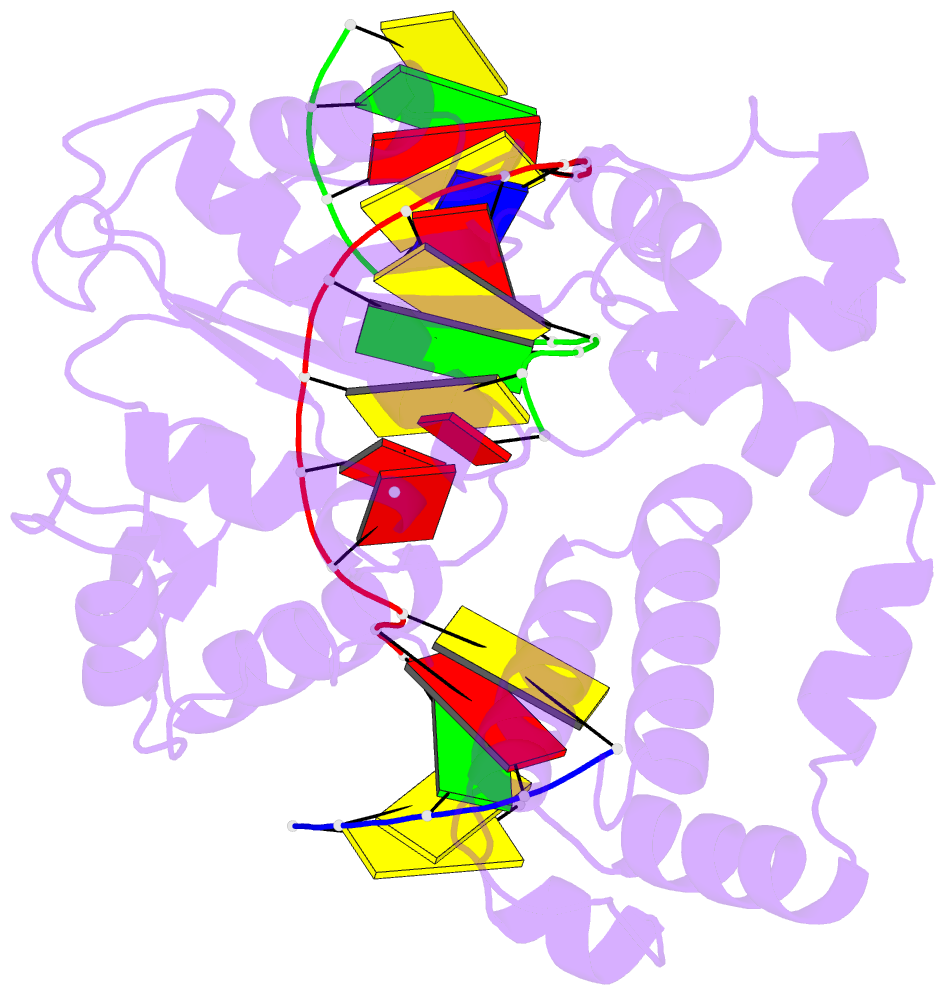
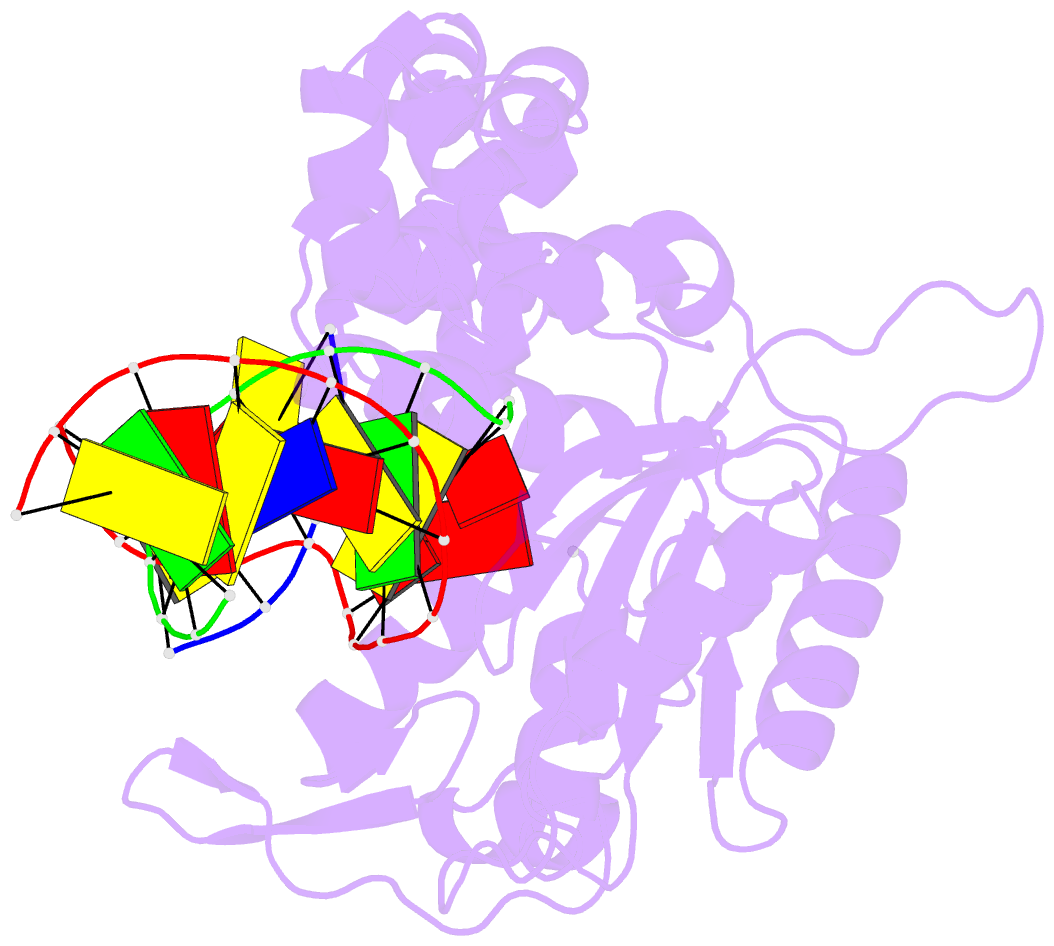
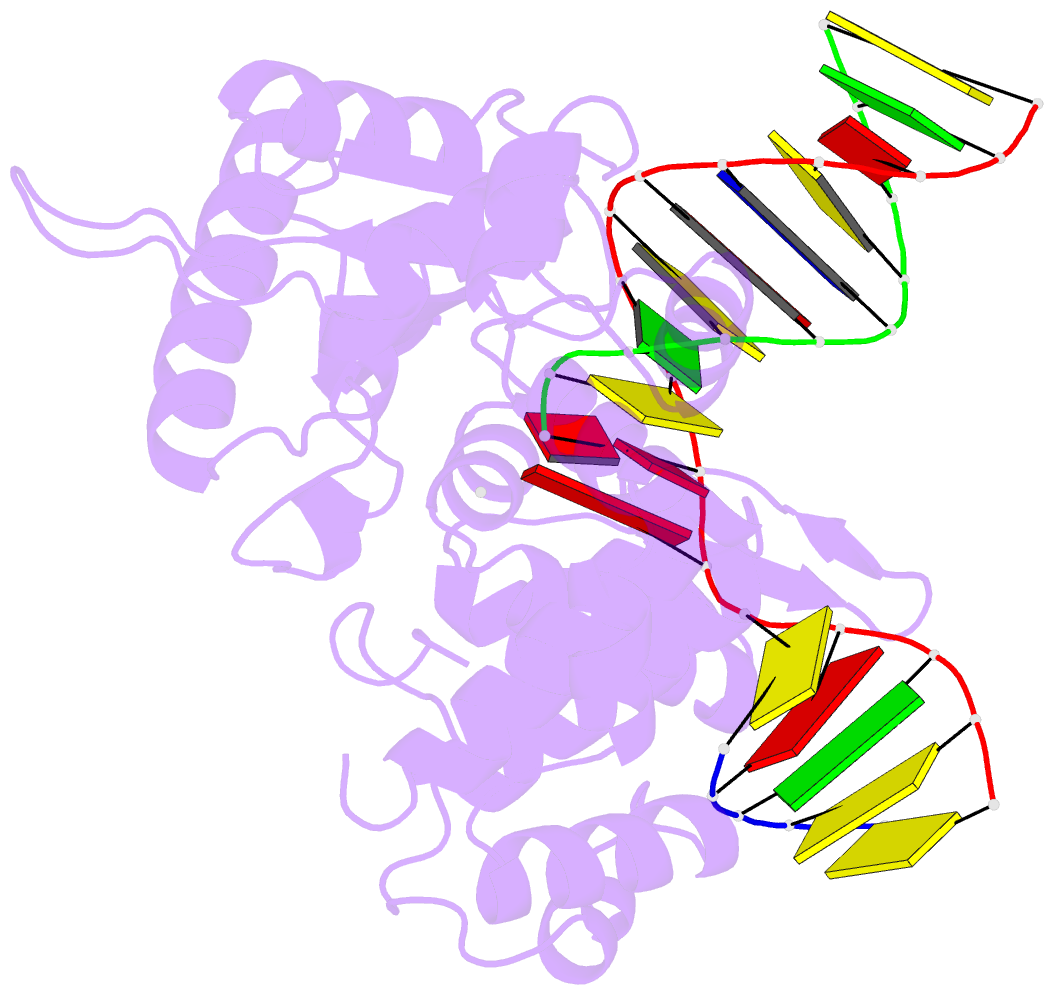
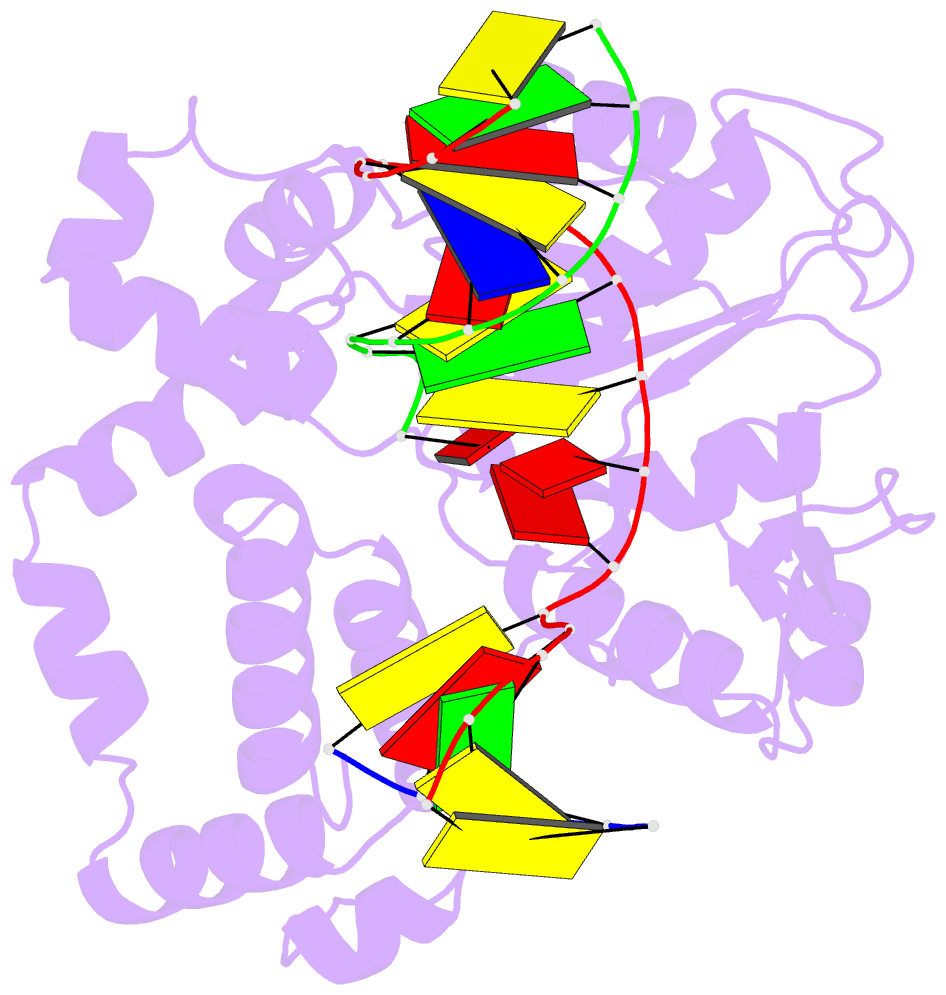
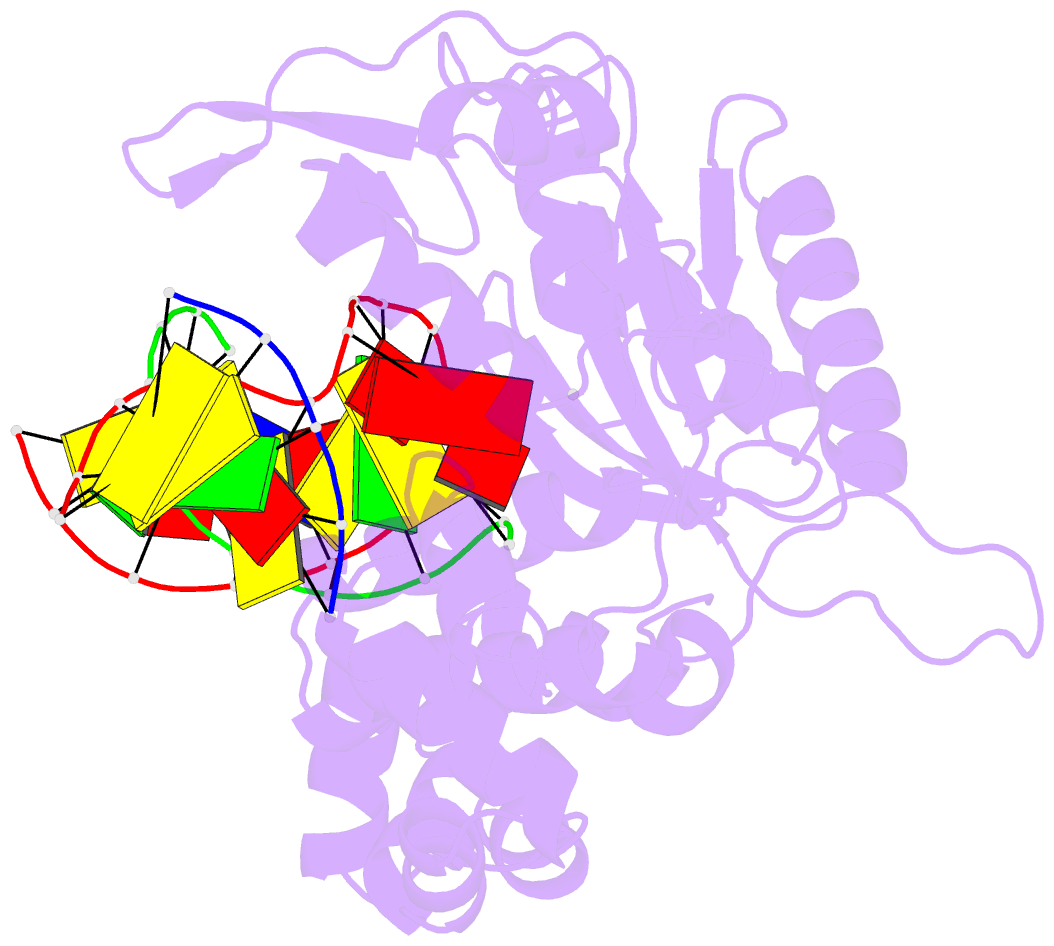
Summary information and primary citation
- PDB-id
- 5j29; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.2 Å)
- Summary
- Ternary complex crystal structure of DNA polymerase beta with a:a mismatch at the primer terminus
- Reference
- Batra VK, Beard WA, Pedersen LC, Wilson SH (2016): "Structures of DNA Polymerase Mispaired DNA Termini Transitioning to Pre-catalytic Complexes Support an Induced-Fit Fidelity Mechanism." Structure, 24, 1863-1875. doi: 10.1016/j.str.2016.08.006.
- Abstract
- High-fidelity DNA synthesis requires that polymerases display a strong preference for right nucleotide insertion. When the wrong nucleotide is inserted, the polymerase deters extension from the mismatched DNA terminus. Twenty-three crystallographic structures of DNA polymerase β with terminal template-primer mismatches were determined as binary DNA and ternary pre-catalytic substrate complexes. These structures indicate that the mismatched termini adopt various distorted conformations that attempt to satisfy stacking and hydrogen-bonding interactions. The binary complex structures indicate an induced strain in the mismatched template nucleotide. Addition of a non-hydrolyzable incoming nucleotide stabilizes the templating nucleotide with concomitant strain in the primer terminus. Several dead-end ternary complex structures suggest that DNA synthesis might occur as the enzyme transitions from an open to a closed complex. The structures are consistent with an induced-fit mechanism where a mismatched terminus is misaligned relative to the correct incoming nucleotide to deter or delay further DNA synthesis.