Summary information and primary citation
- PDB-id
- 5j37; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- virus-DNA
- Method
- X-ray (2.3 Å)
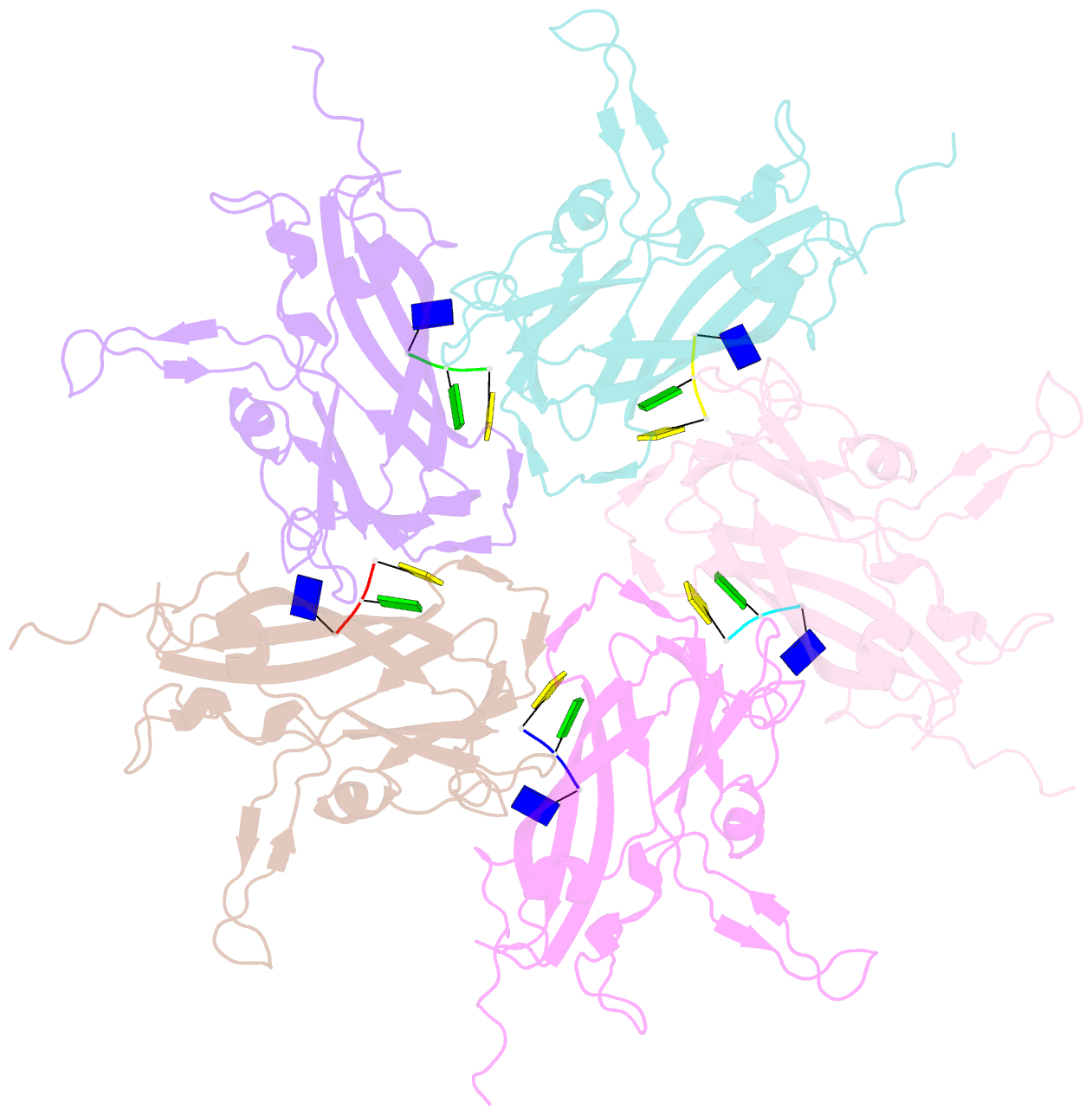
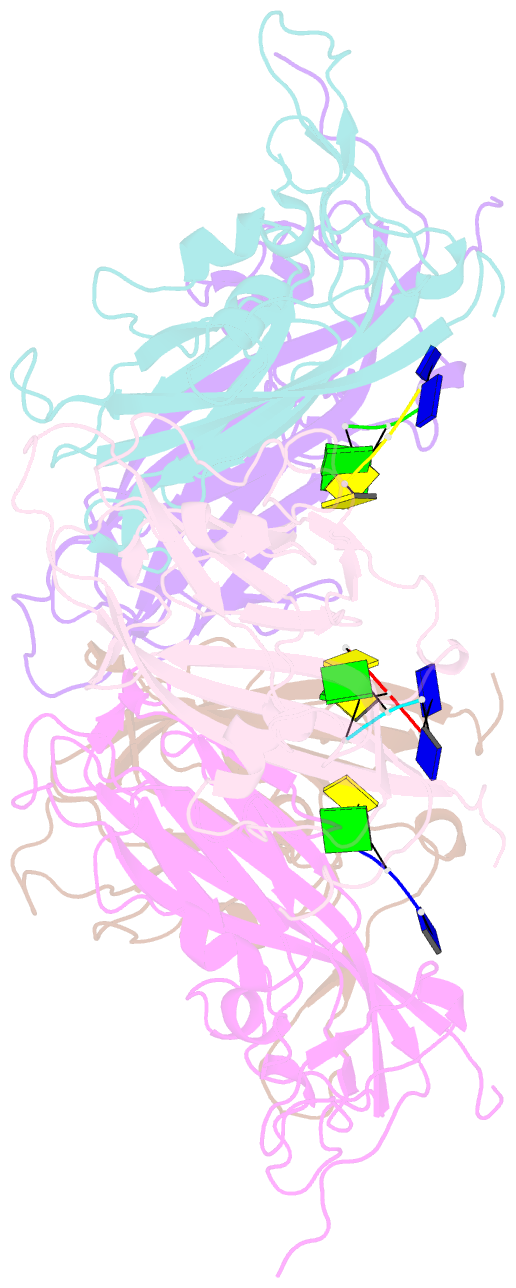
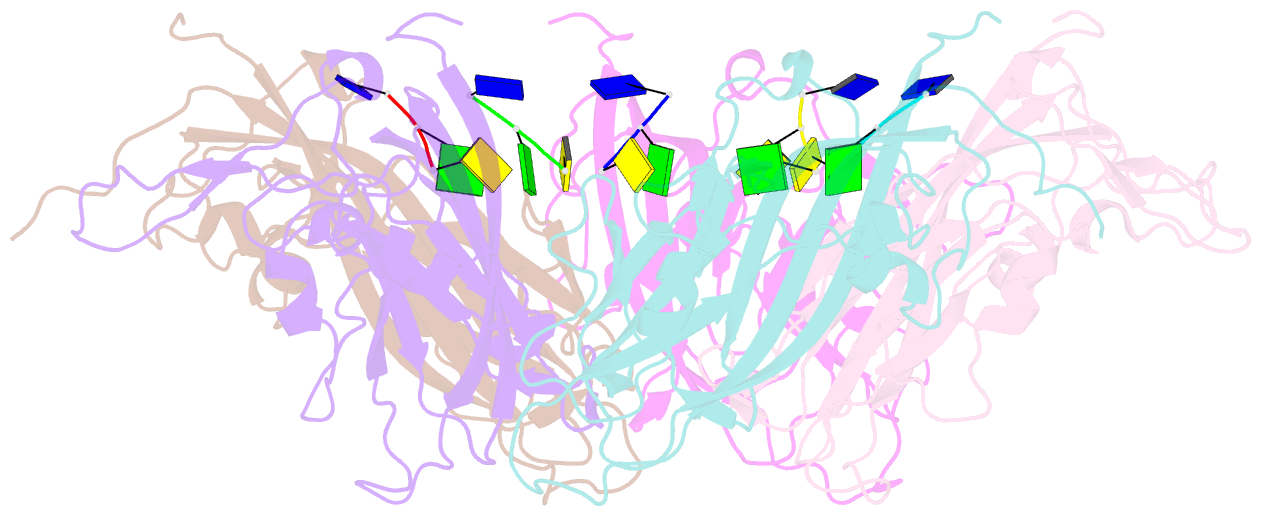
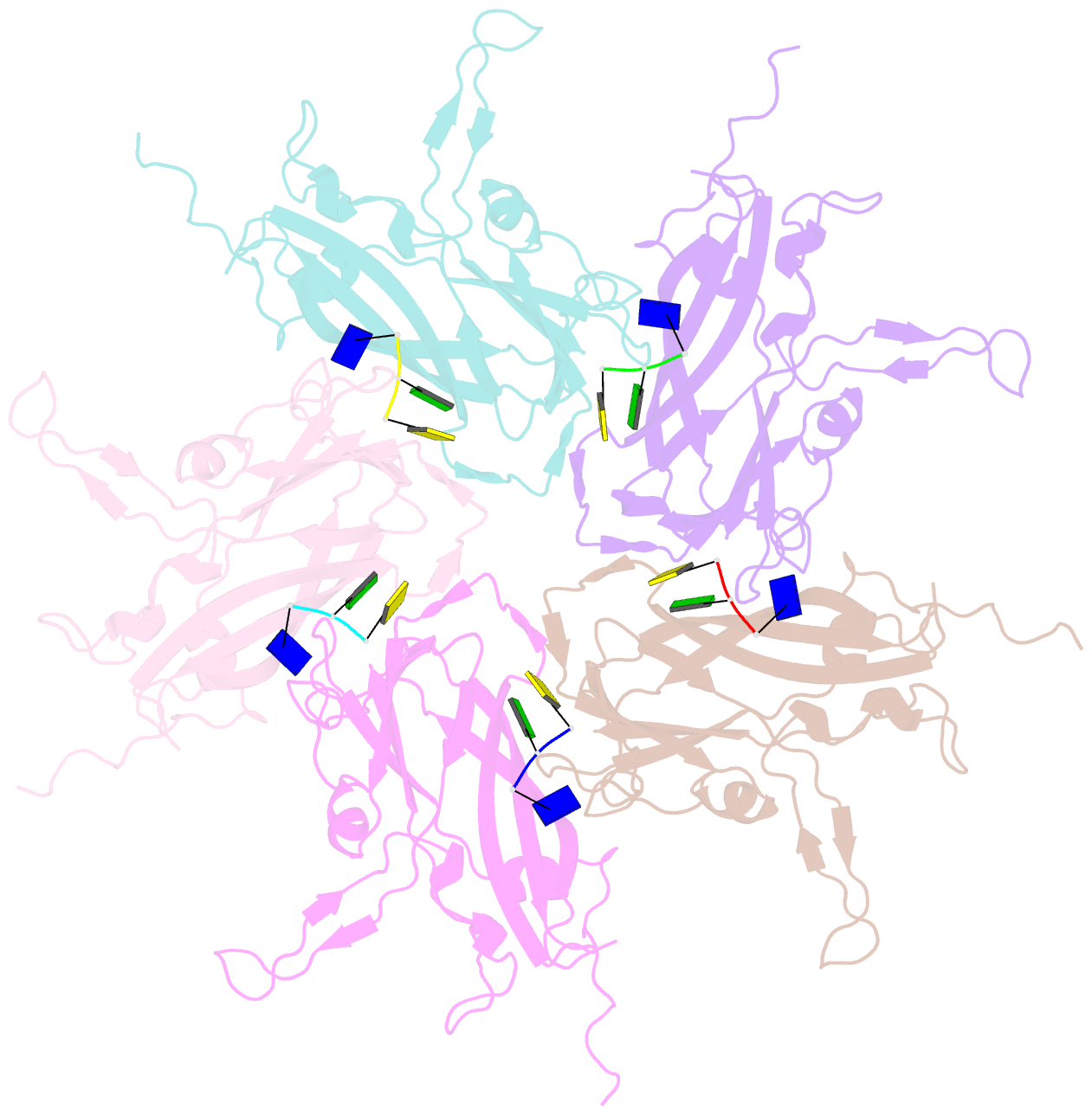
- Summary
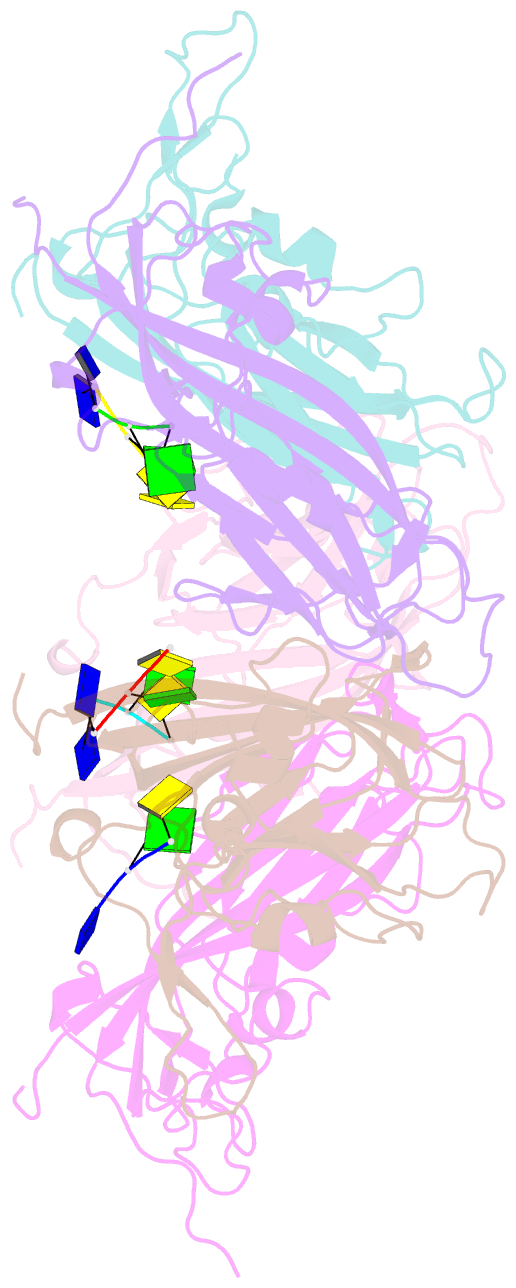
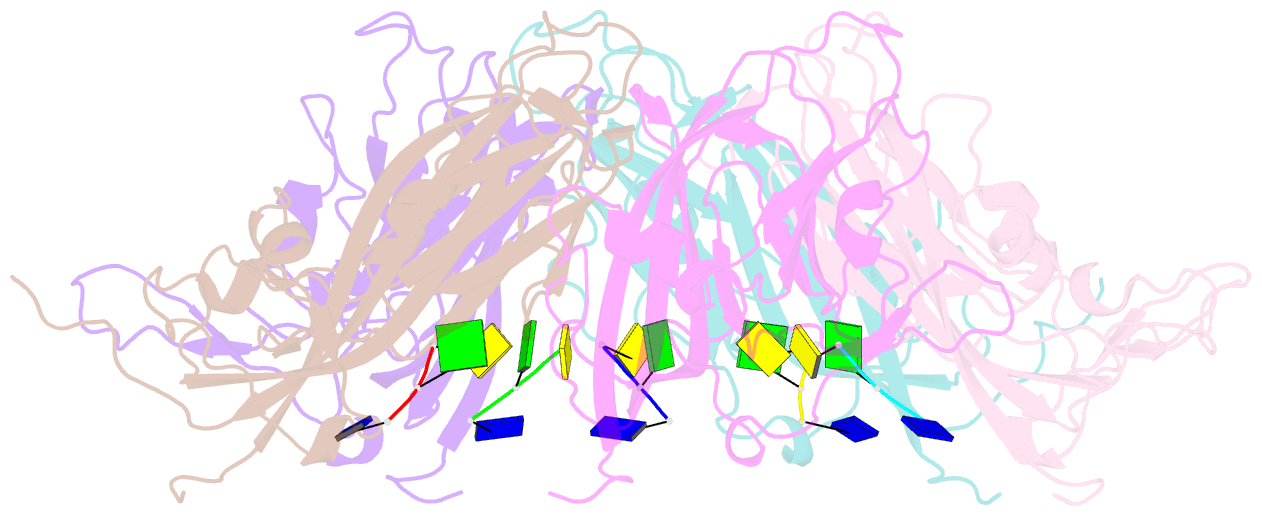
- Crystal structure of 60-mer bfdv capsid protein in complex with single stranded DNA
- Reference
- Sarker S, Terron MC, Khandokar Y, Aragao D, Hardy JM, Radjainia M, Jimenez-Zaragoza M, de Pablo PJ, Coulibaly F, Luque D, Raidal SR, Forwood JK (2016): "Structural insights into the assembly and regulation of distinct viral capsid complexes." Nat Commun, 7, 13014. doi: 10.1038/ncomms13014.
- Abstract
- The assembly and regulation of viral capsid proteins into highly ordered macromolecular complexes is essential for viral replication. Here, we utilize crystal structures of the capsid protein from the smallest and simplest known viruses capable of autonomously replicating in animal cells, circoviruses, to establish structural and mechanistic insights into capsid morphogenesis and regulation. The beak and feather disease virus, like many circoviruses, encode only two genes: a capsid protein and a replication initiation protein. The capsid protein forms distinct macromolecular assemblies during replication and here we elucidate these structures at high resolution, showing that these complexes reverse the exposure of the N-terminal arginine rich domain responsible for DNA binding and nuclear localization. We show that assembly of these complexes is regulated by single-stranded DNA (ssDNA), and provide a structural basis of capsid assembly around single-stranded DNA, highlighting novel binding interfaces distinct from the highly positively charged N-terminal ARM domain.