Summary information and primary citation
- PDB-id
- 5jk0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- cell cycle
- Method
- X-ray (2.1 Å)
- Summary
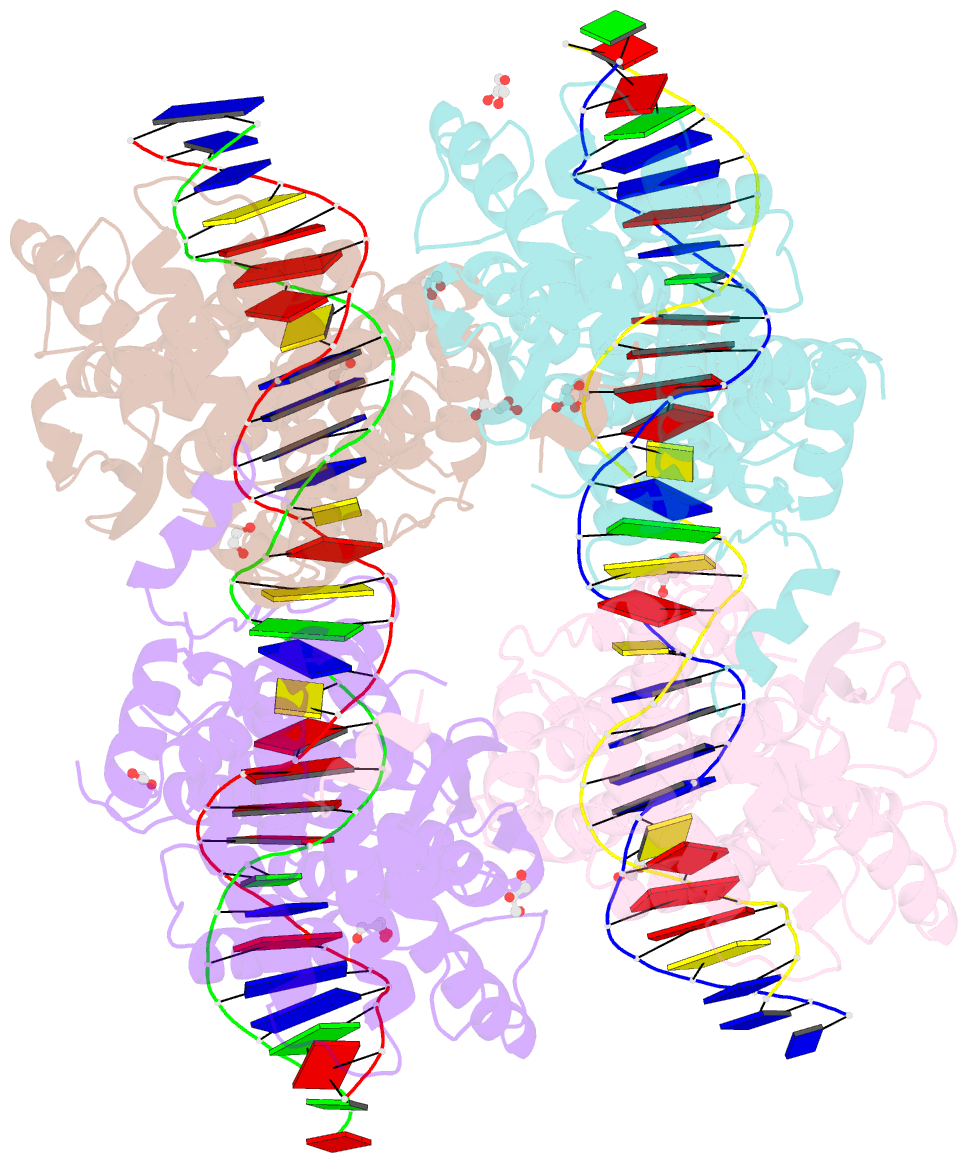
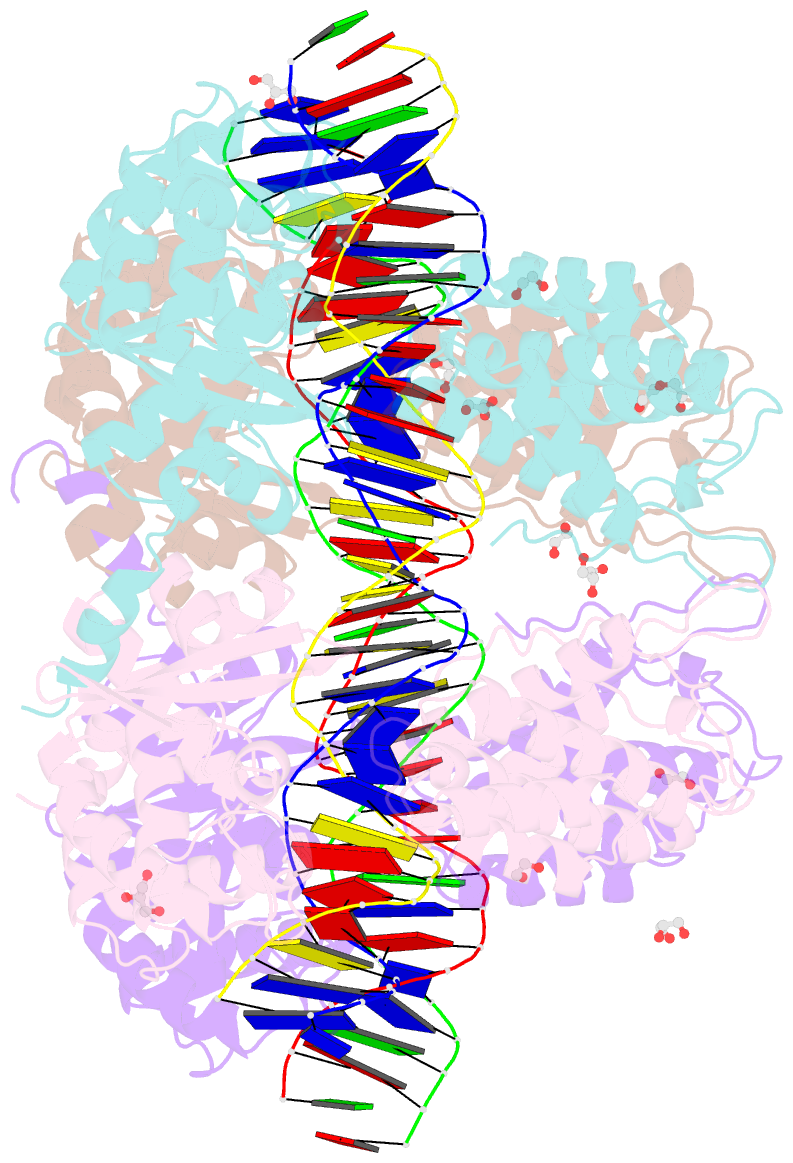
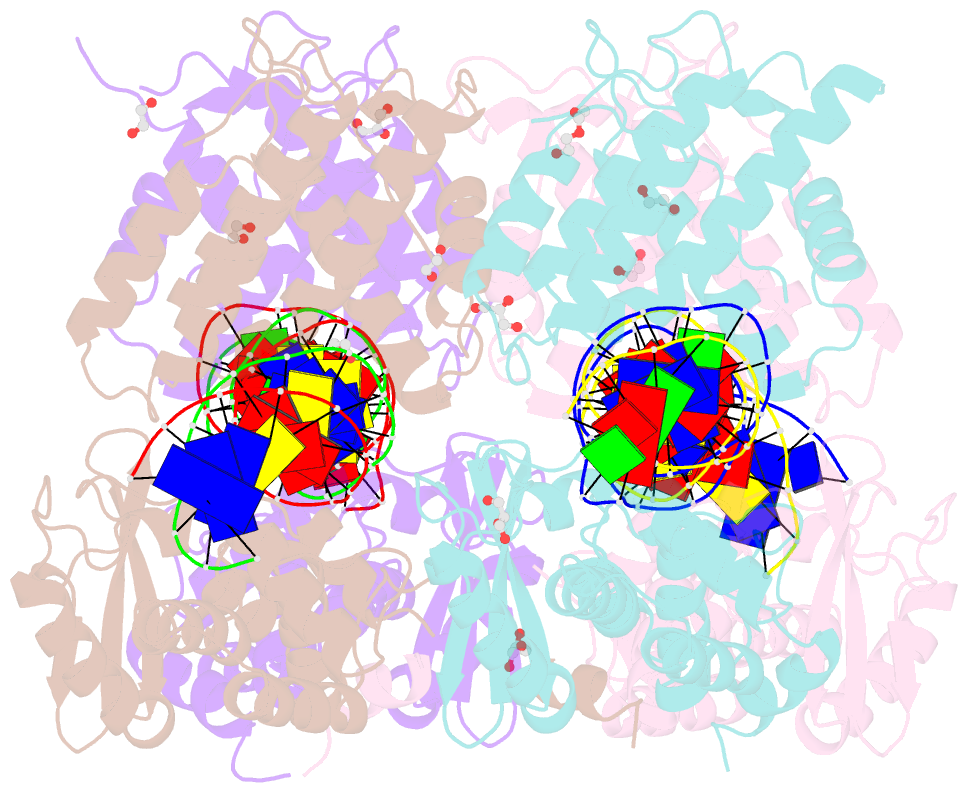
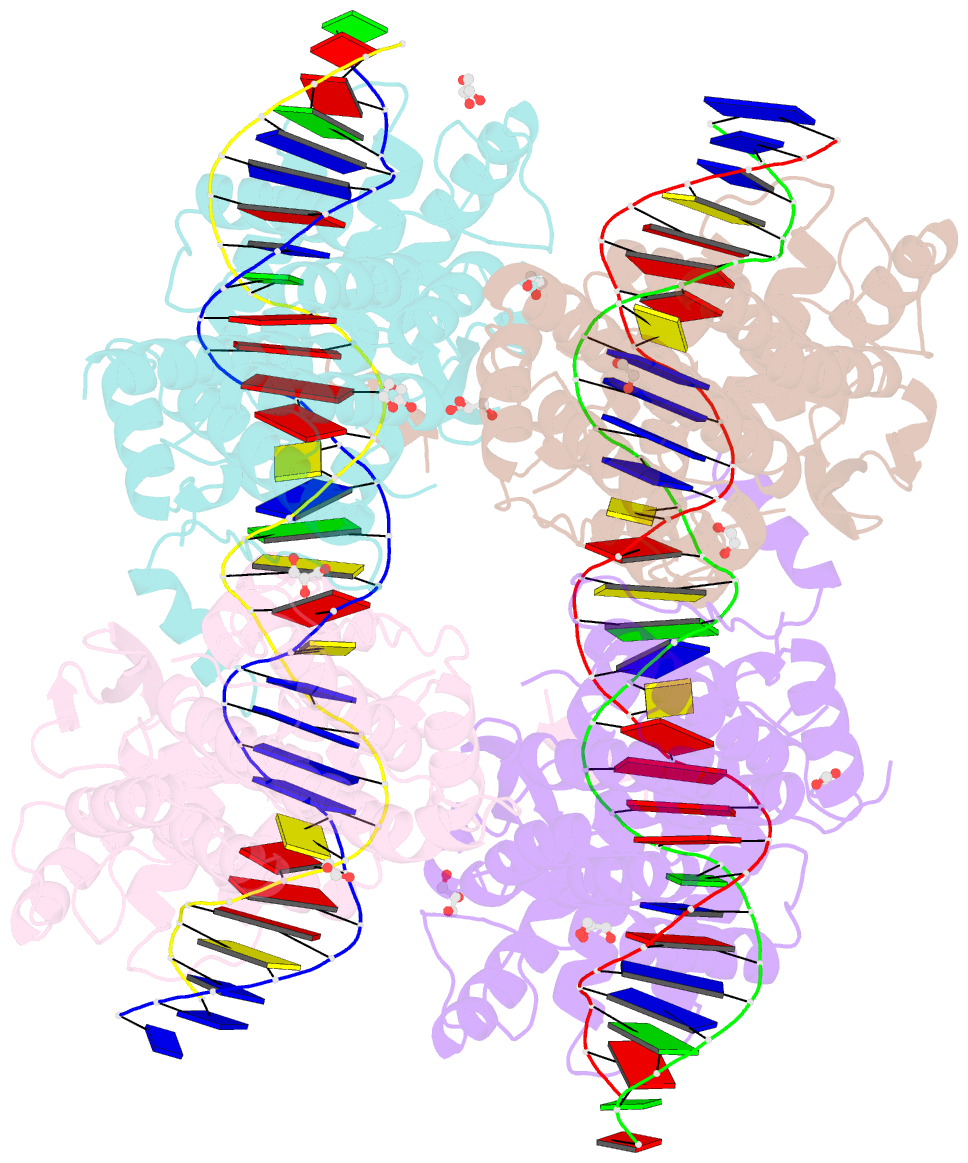
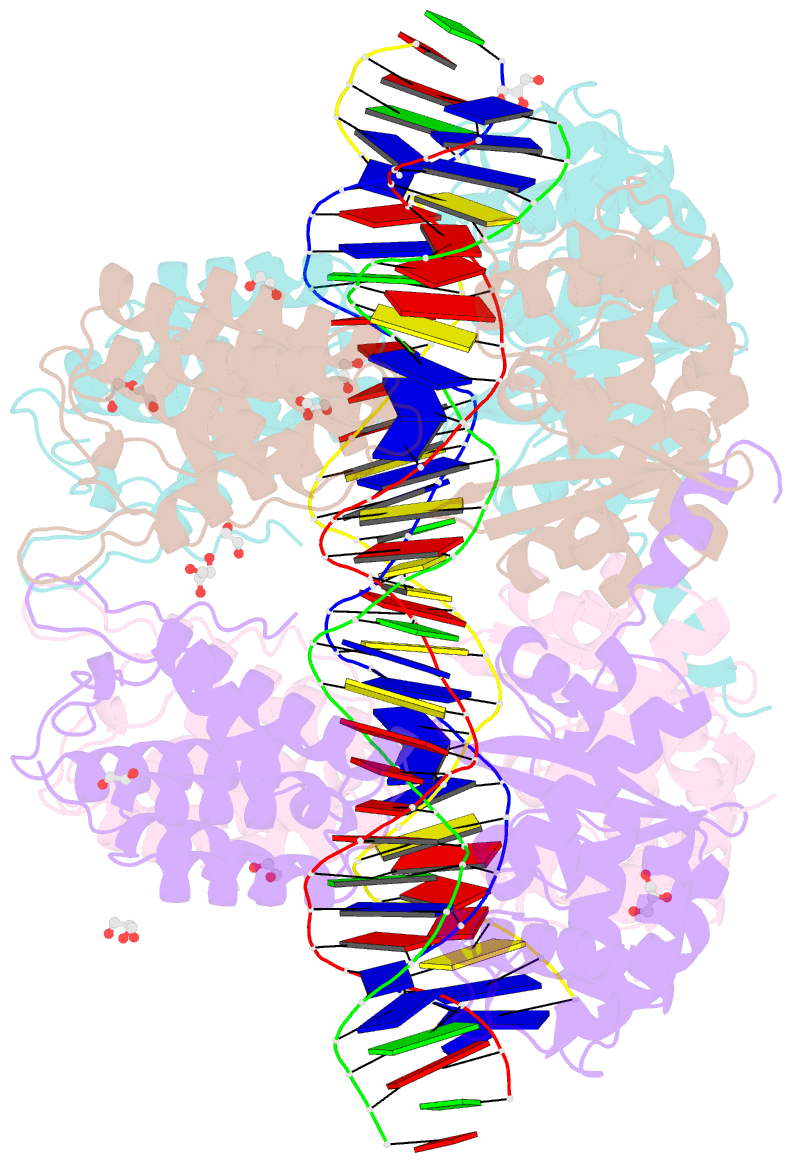
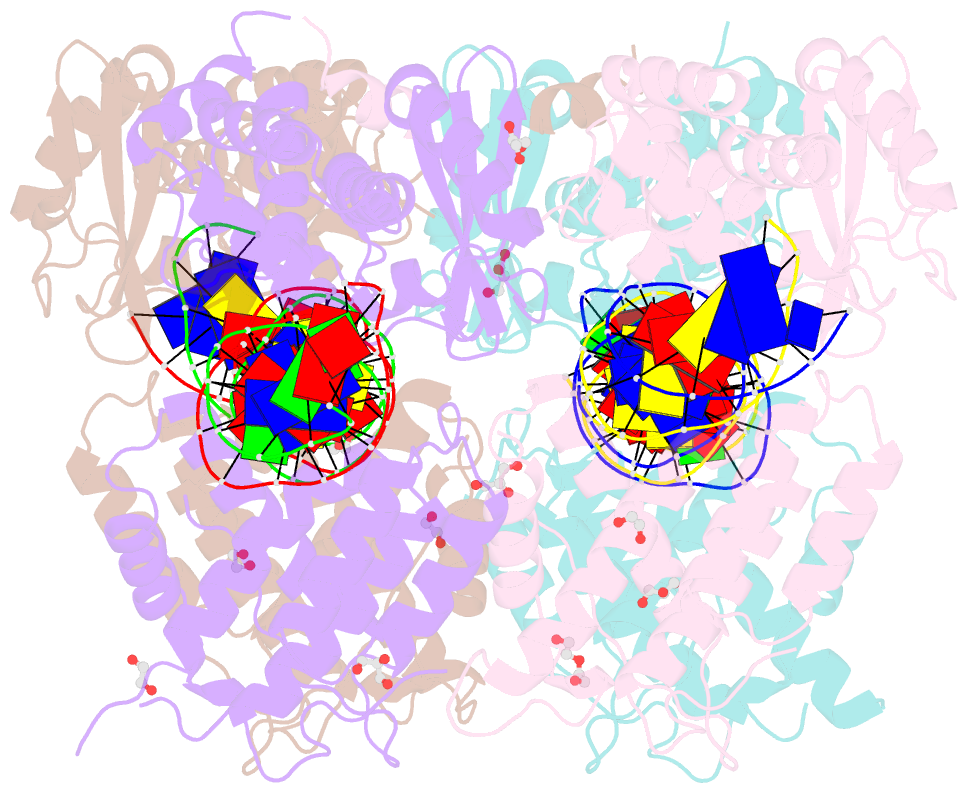
- Crystal structure of xerh site-specific recombinase bound to difh substrate: pre-cleavage complex
- Reference
- Bebel A, Karaca E, Kumar B, Stark WM, Barabas O (2016): "Structural snapshots of Xer recombination reveal activation by synaptic complex remodeling and DNA bending." Elife, 5. doi: 10.7554/eLife.19706.
- Abstract
- Bacterial Xer site-specific recombinases play an essential genome maintenance role by unlinking chromosome multimers, but their mechanism of action has remained structurally uncharacterized. Here, we present two high-resolution structures of Helicobacter pylori XerH with its recombination site DNA difH, representing pre-cleavage and post-cleavage synaptic intermediates in the recombination pathway. The structures reveal that activation of DNA strand cleavage and rejoining involves large conformational changes and DNA bending, suggesting how interaction with the cell division protein FtsK may license recombination at the septum. Together with biochemical and in vivo analysis, our structures also reveal how a small sequence asymmetry in difH defines protein conformation in the synaptic complex and orchestrates the order of DNA strand exchanges. Our results provide insights into the catalytic mechanism of Xer recombination and a model for regulation of recombination activity during cell division.