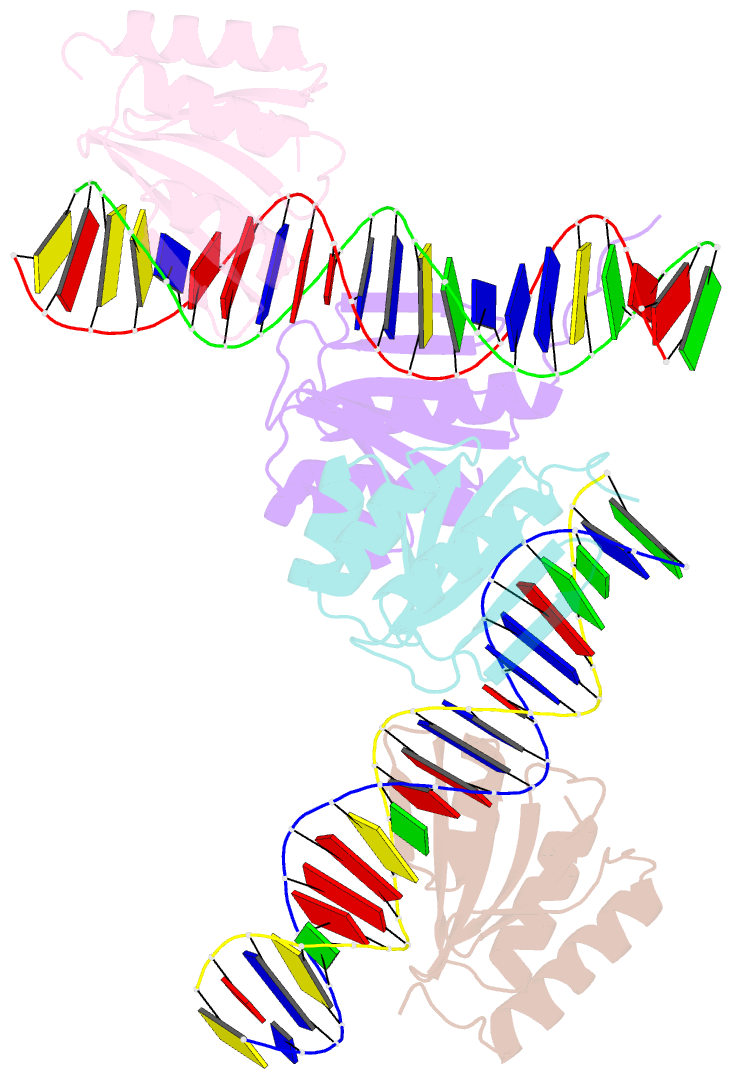
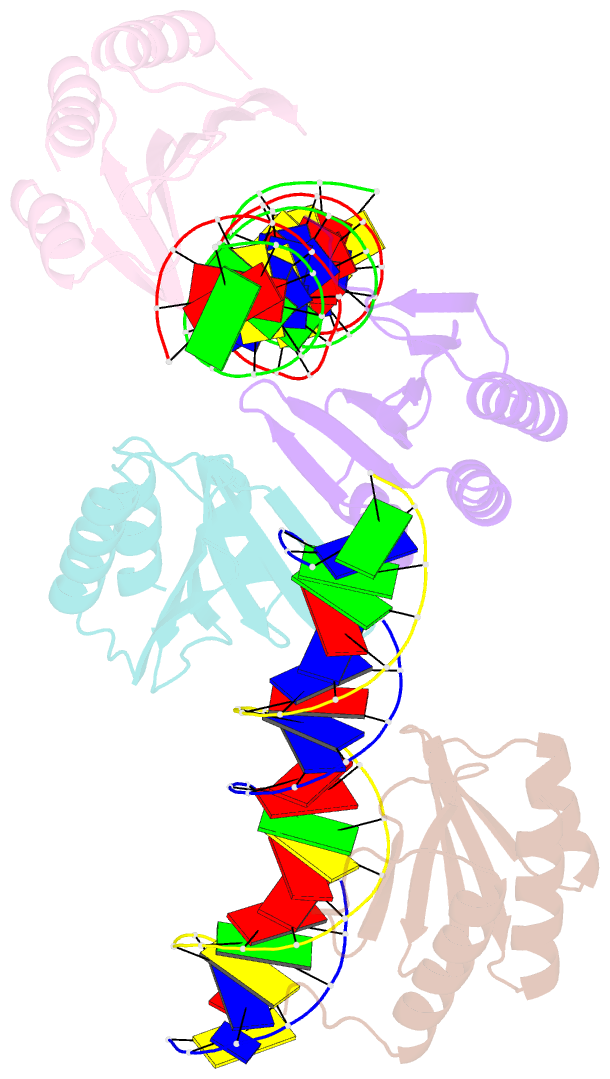
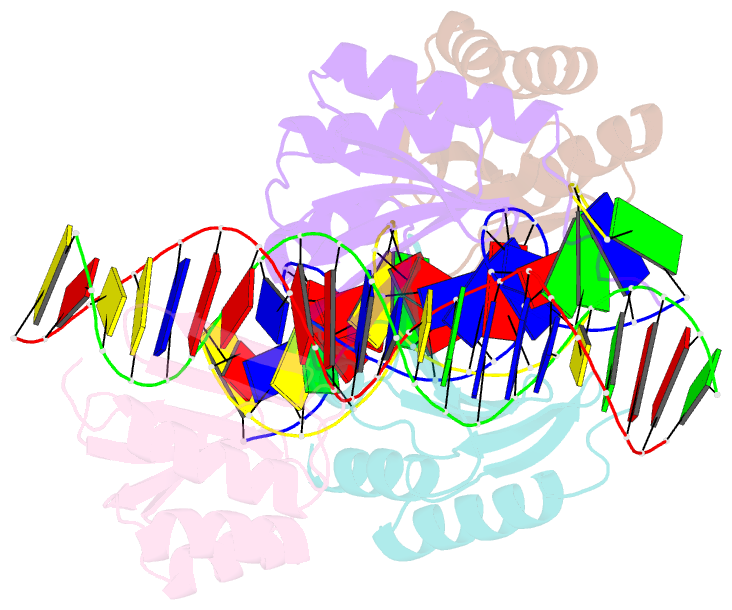
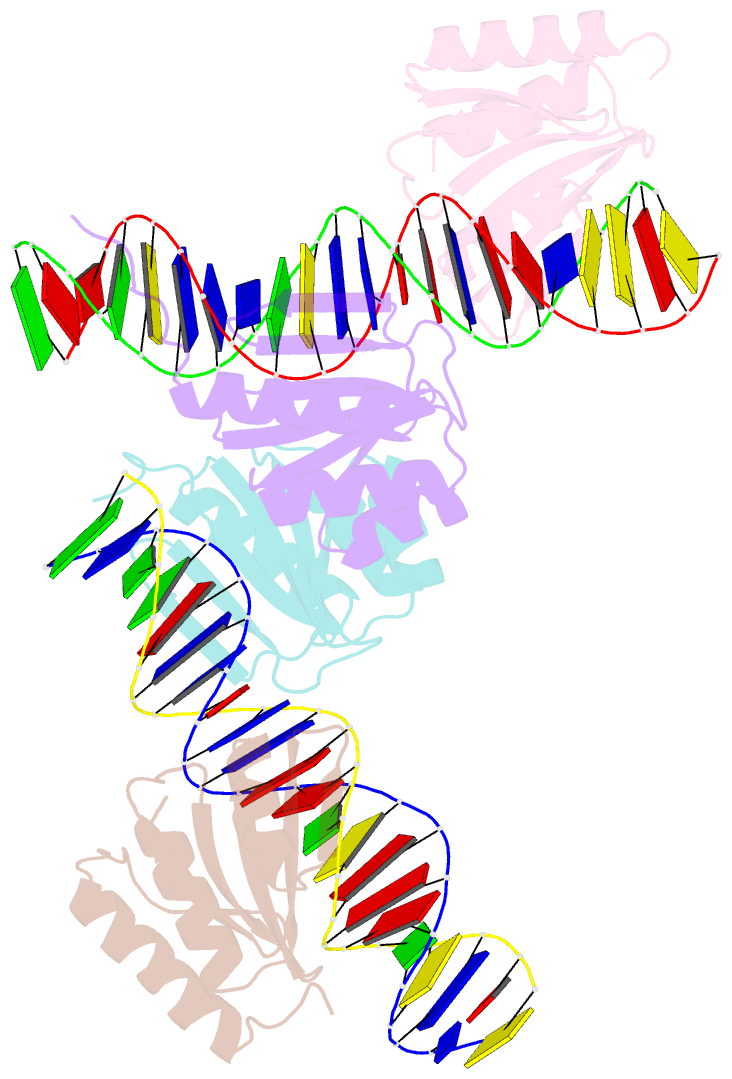
Summary information and primary citation
- PDB-id
- 5jlt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA
- Method
- X-ray (2.955 Å)
- Summary
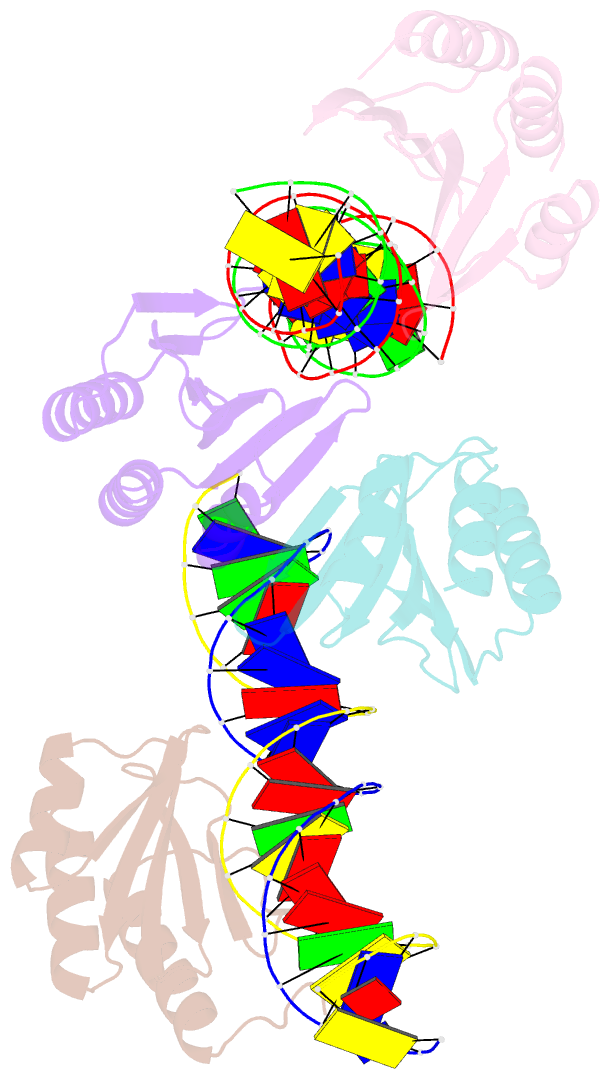
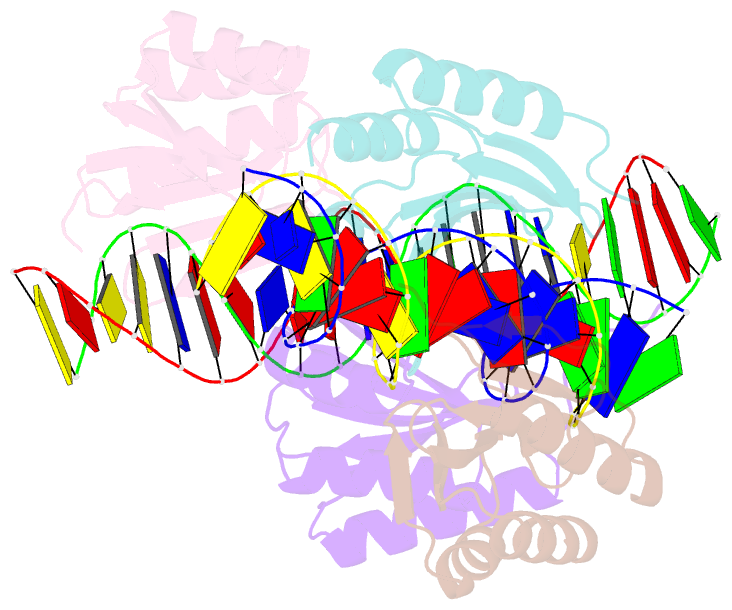
- The crystal structure of the bacteriophage t4 mota c-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif
- Reference
- Cuypers MG, Robertson RM, Knipling L, Waddell MB, Moon K, Hinton DM, White SW (2018): "The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA." Nucleic Acids Res., 46, 5308-5318. doi: 10.1093/nar/gky292.
- Abstract
- During infection, bacteriophage T4 produces the MotA transcription factor that redirects the host RNA polymerase to the expression of T4 middle genes. The C-terminal 'double-wing' domain of MotA binds specifically to the MotA box motif of middle T4 promoters. We report the crystal structure of this complex, which reveals a new mode of protein-DNA interaction. The domain binds DNA mostly via interactions with the DNA backbone, but the binding is enhanced in the specific cognate structure by additional interactions with the MotA box motif in both the major and minor grooves. The linker connecting the two MotA domains plays a key role in stabilizing the complex via minor groove interactions. The structure is consistent with our previous model derived from chemical cleavage experiments using the entire transcription complex. α- and β-d-glucosyl-5-hydroxymethyl-deoxycytosine replace cytosine in T4 DNA, and docking simulations indicate that a cavity in the cognate structure can accommodate the modified cytosine. Binding studies confirm that the modification significantly enhances the binding affinity of MotA for the DNA. Consequently, our work reveals how a DNA modification can extend the uniqueness of small DNA motifs to facilitate the specificity of protein-DNA interactions.