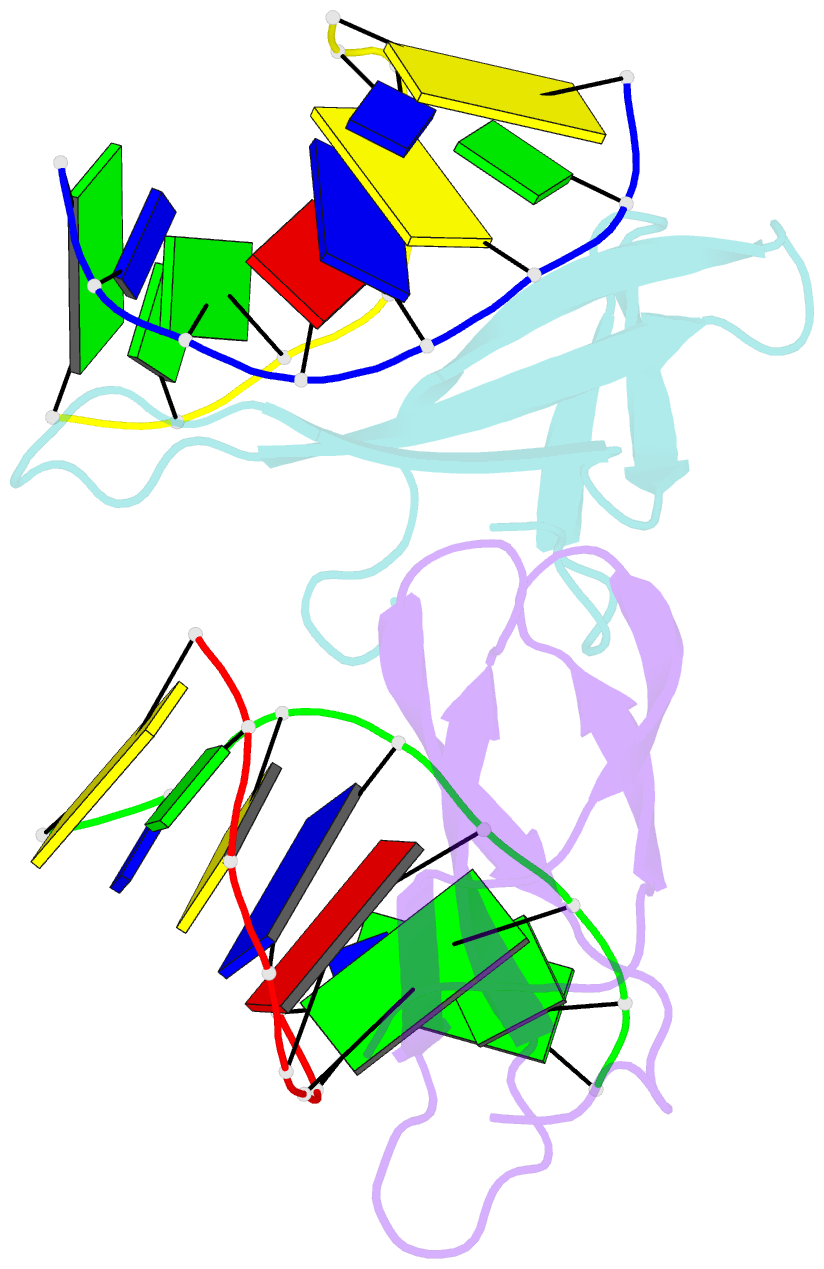
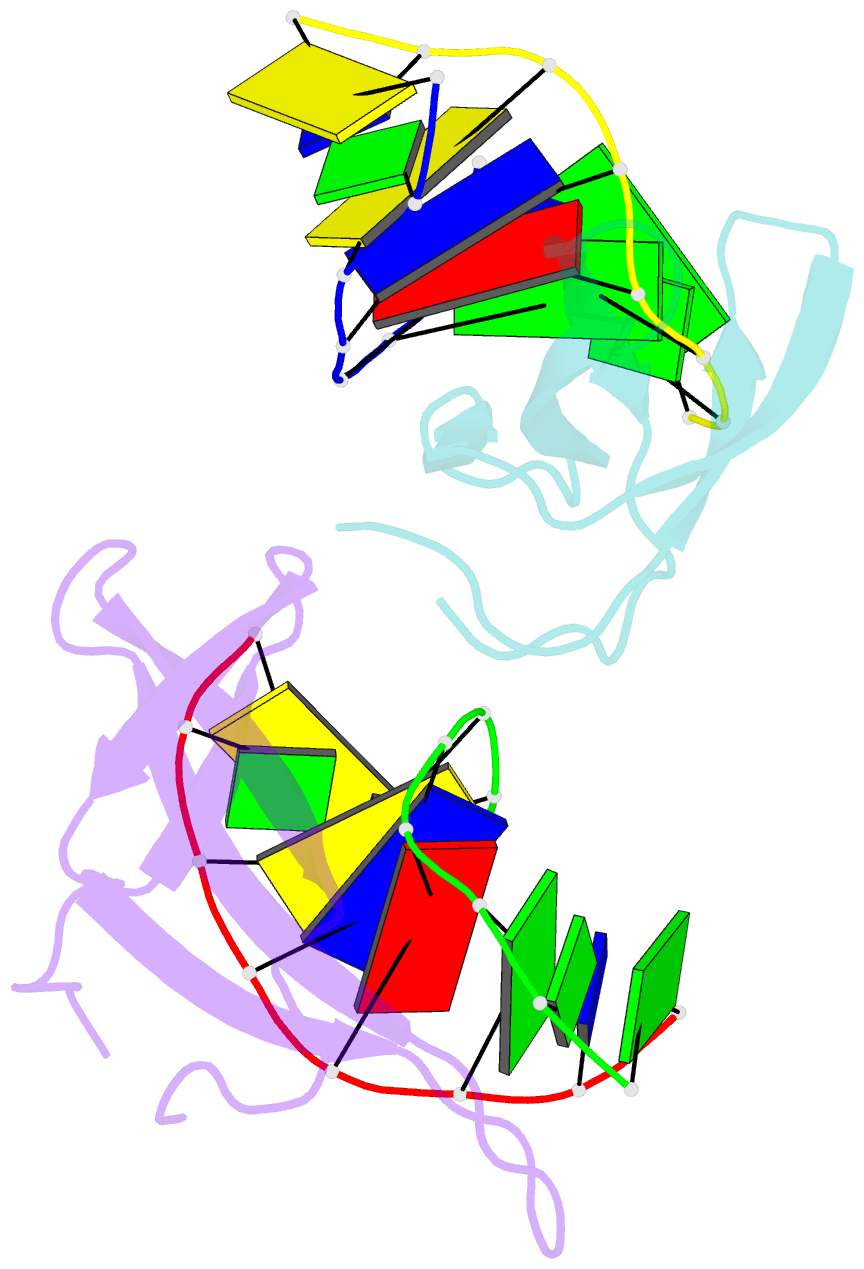
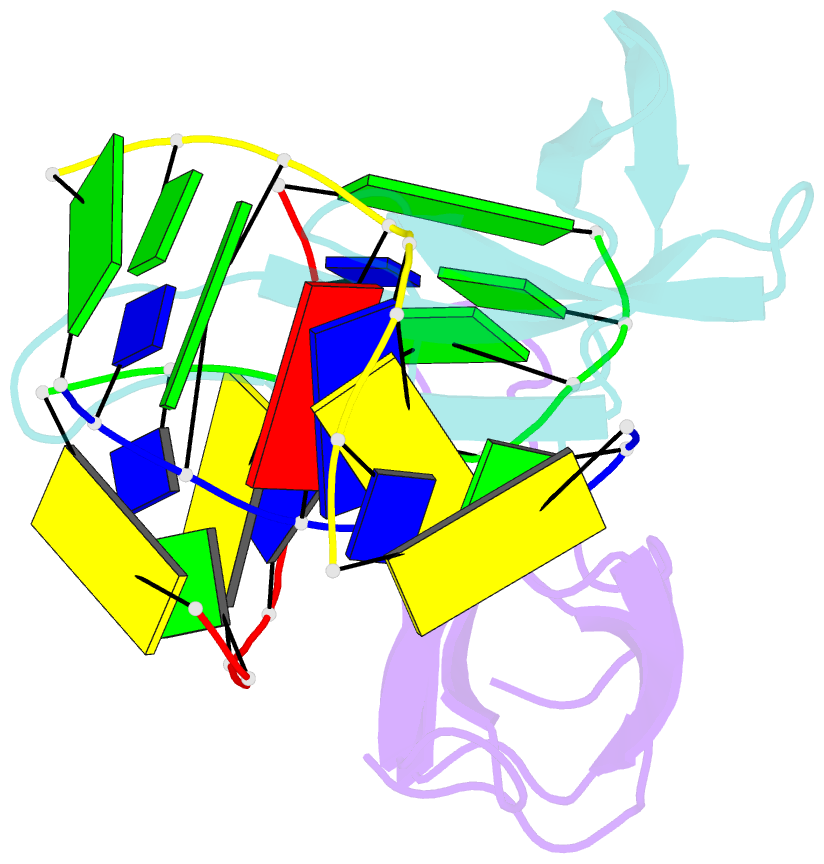
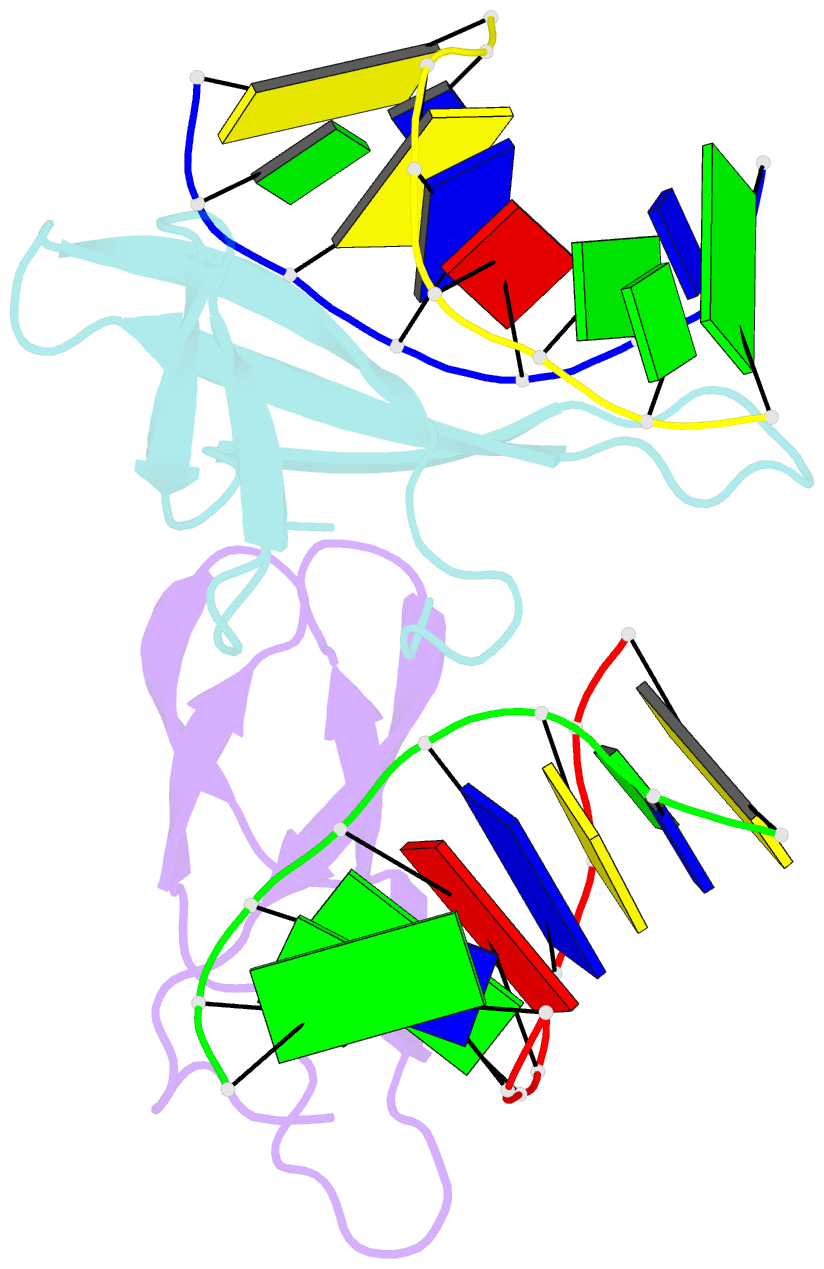
Summary information and primary citation
- PDB-id
- 5k17; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.1 Å)
- Summary
- Crystal structure of cren7-dsDNA (gtgatcgc) complex
- Reference
- Tian L, Zhang ZF, Wang H, Zhao M, Dong Y, Gong Y (2016): "Sequence-Dependent T:G Base Pair Opening in DNA Double Helix Bound by Cren7, a Chromatin Protein Conserved among Crenarchaea." PLoS ONE, 11, e0163361. doi: 10.1371/journal.pone.0163361.
- Abstract
- T:G base pair arising from spontaneous deamination of 5mC or polymerase errors is a great challenge for DNA repair of hyperthermophilic archaea, especially Crenarchaea. Most strains in this phylum lack the protein homologues responsible for the recognition of the mismatch in the DNA repair pathways. To investigate whether Cren7, a highly conserved chromatin protein in Crenarchaea, serves a role in the repair of T:G mispairs, the crystal structures of Cren7-GTAATTGC and Cren7-GTGATCGC complexes were solved at 2.0 Å and 2.1 Å. In our structures, binding of Cren7 to the AT-rich DNA duplex (GTAATTGC) induces opening of T2:G15 but not T10:G7 base pair. By contrast, both T:G mispairs in the GC-rich DNA duplex (GTGATCGC) retain the classic wobble type. Structural analysis also showed DNA helical changes of GTAATTGC, especially in the steps around the open T:G base pair, as compared to GTGATCGC or the matched DNAs. Surface plasmon resonance assays revealed a 4-fold lower binding affinity of Cren7 for GTAATTGC than that for GTGATCGC, which was dominantly contributed by the decrease of association rate. These results suggested that binding of Cren7 to DNA leads to T:G mispair opening in a sequence dependent manner, and therefore propose the potential roles of Cren7 in DNA repair.