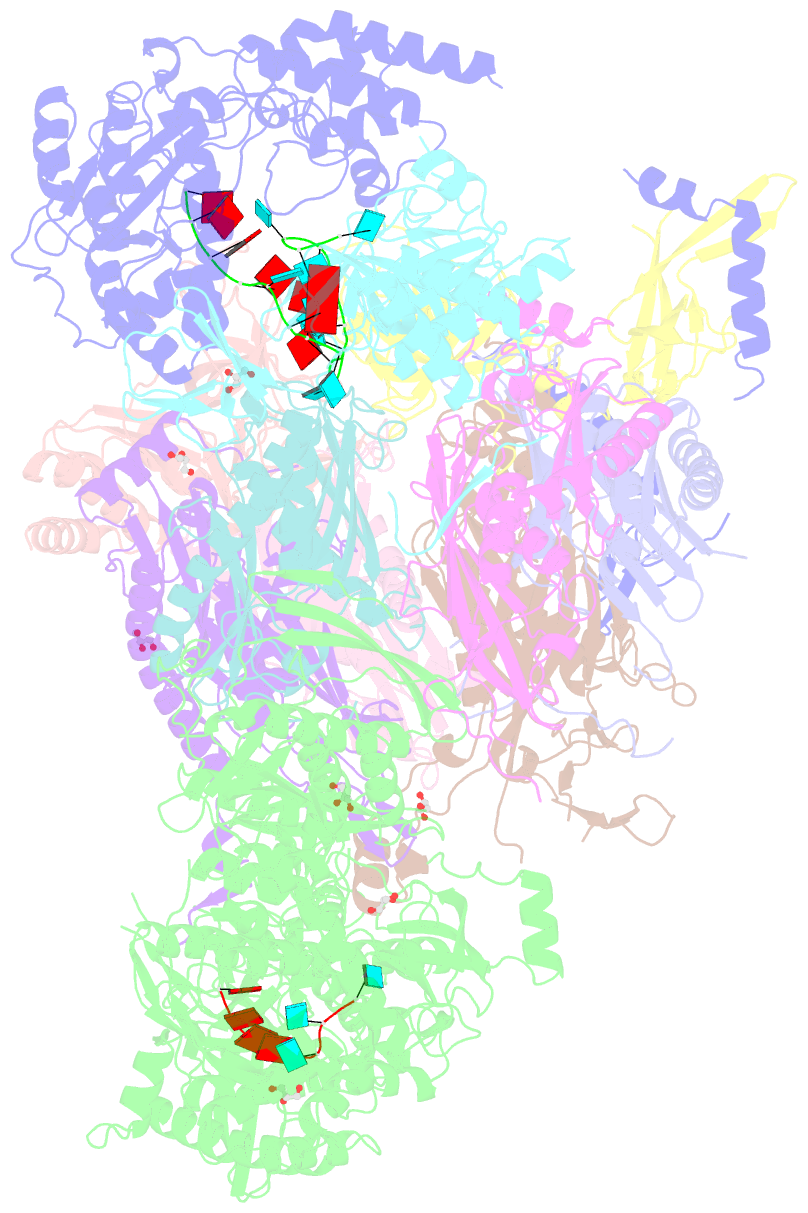
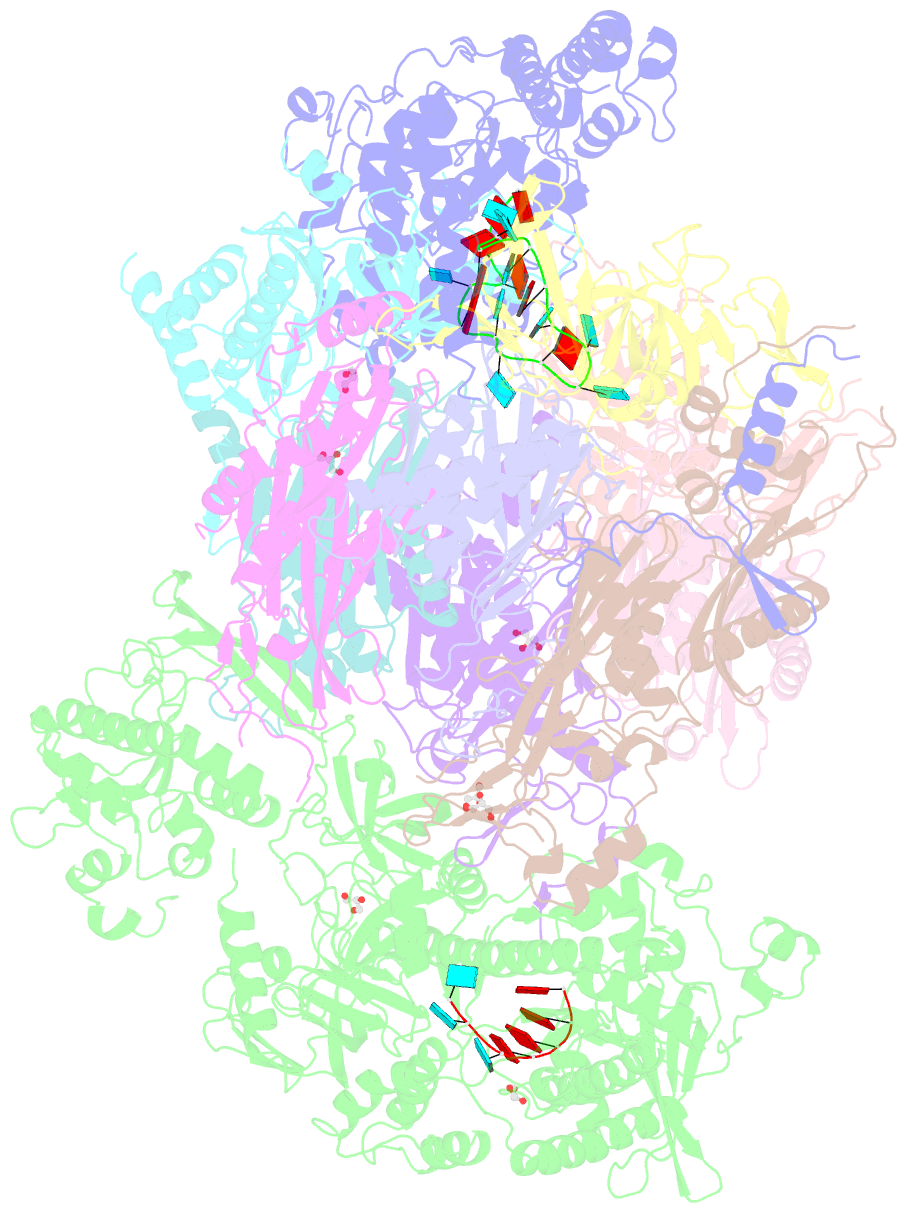
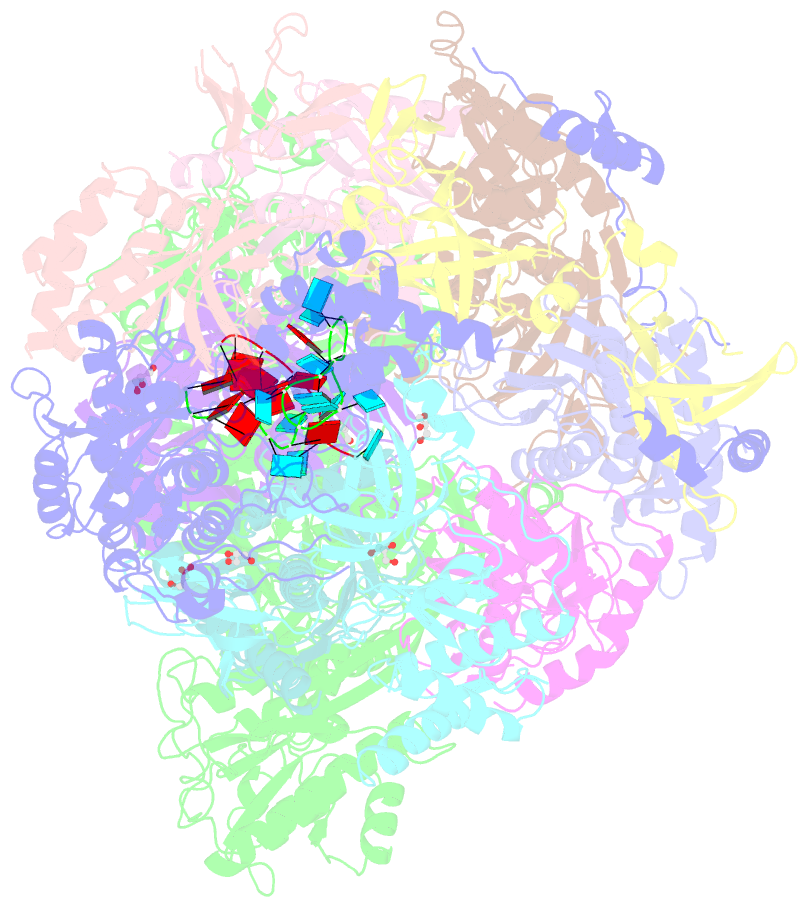
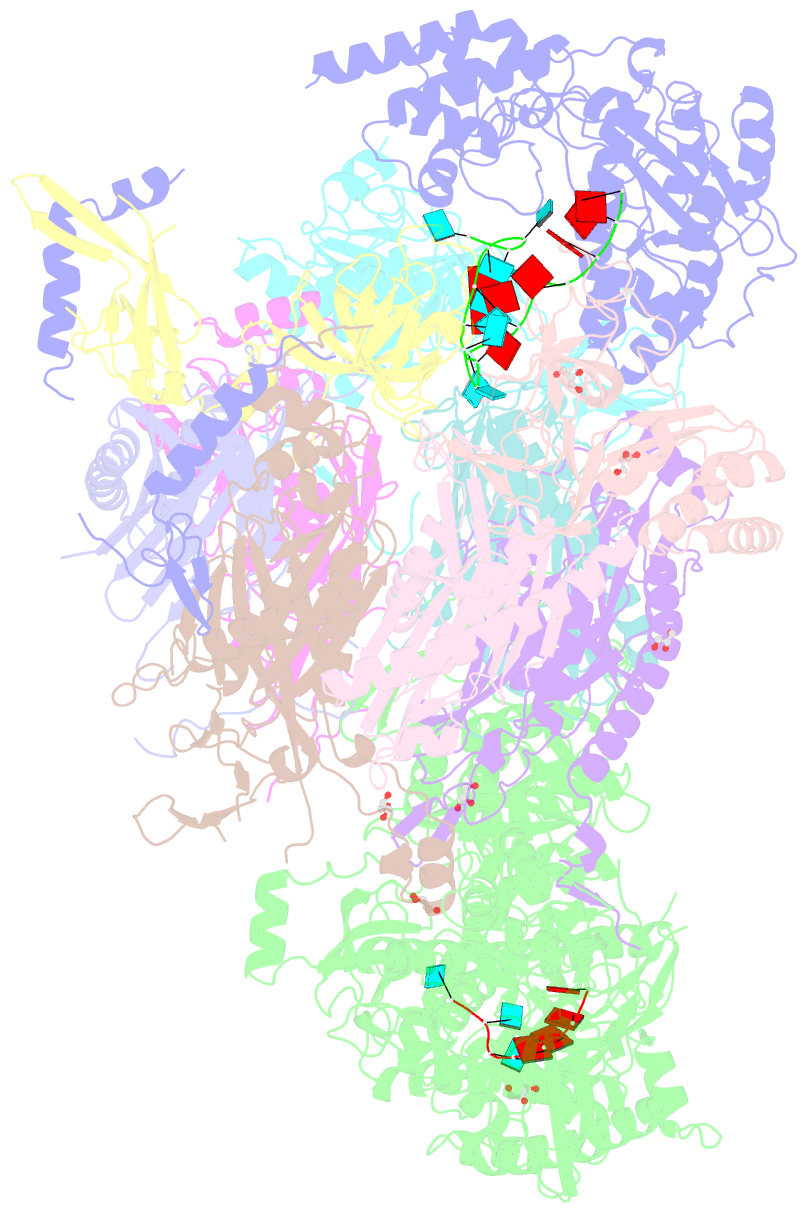
Summary information and primary citation
- PDB-id
- 5k36; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (3.1 Å)
- Summary
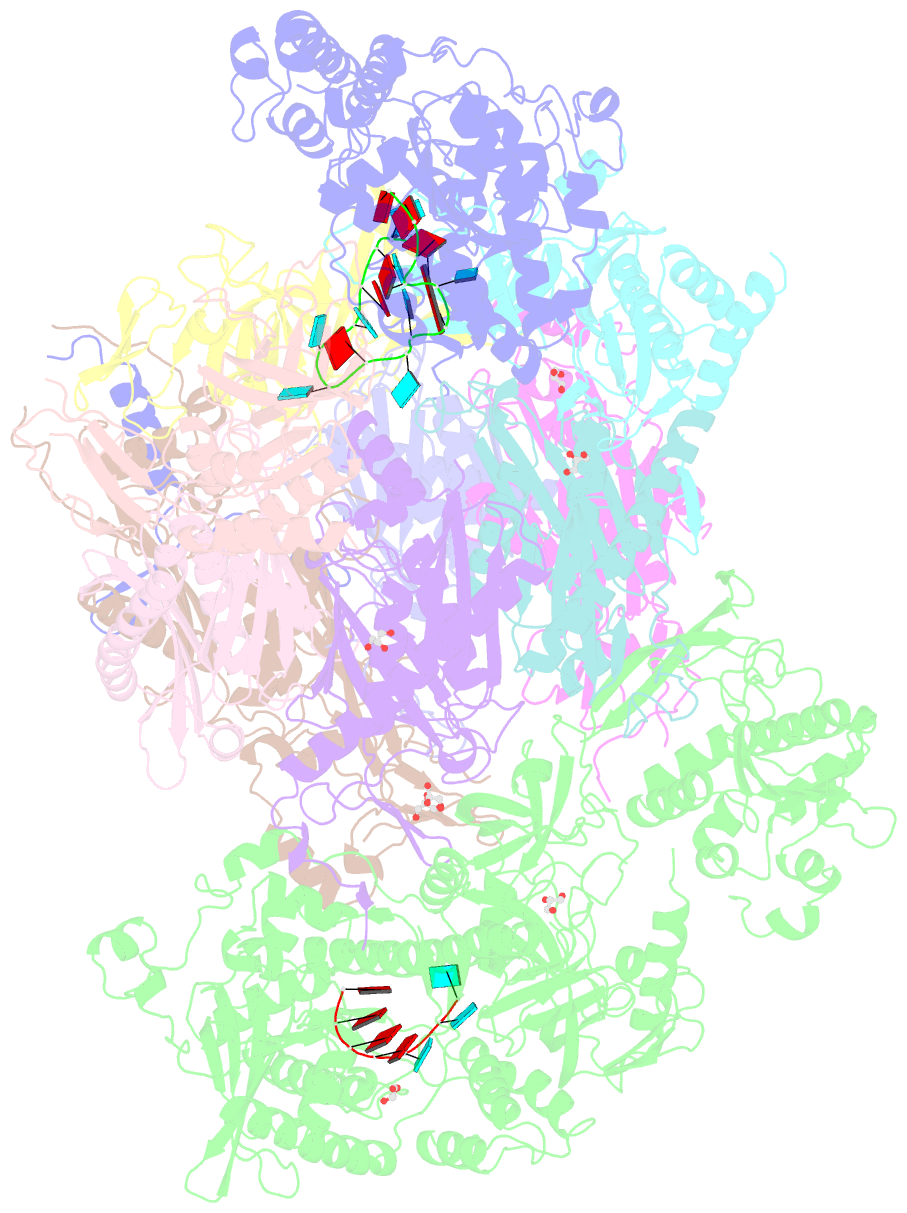
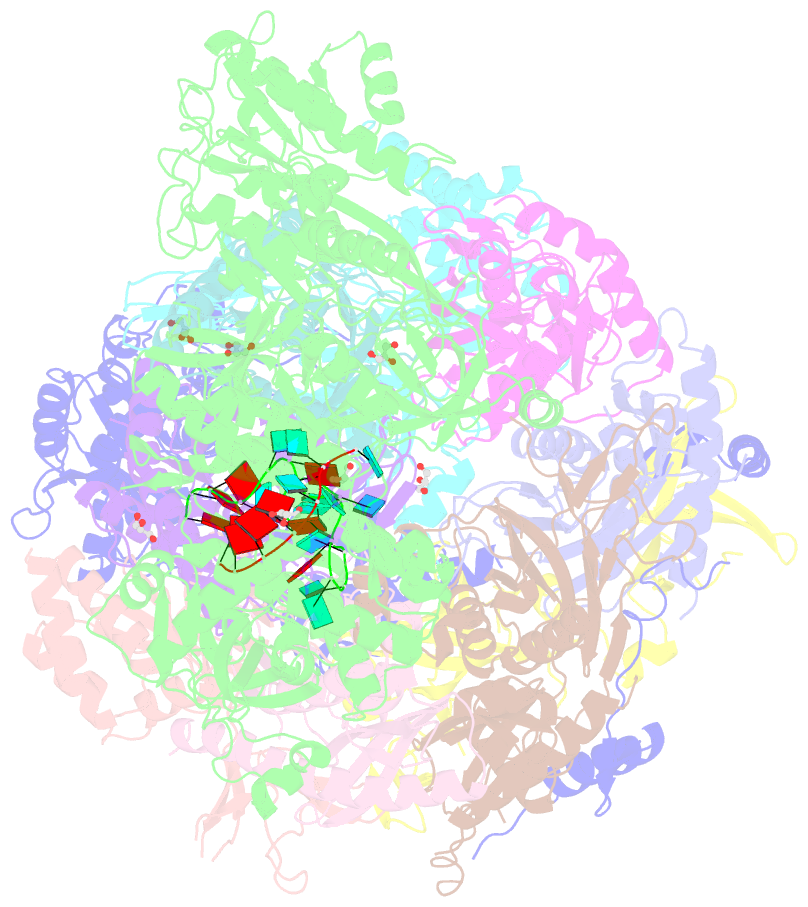
- Structure of an eleven component nuclear RNA exosome complex bound to RNA
- Reference
- Zinder JC, Wasmuth EV, Lima CD (2016): "Nuclear RNA Exosome at 3.1 angstrom Reveals Substrate Specificities, RNA Paths, and Allosteric Inhibition of Rrp44/Dis3." Mol.Cell, 64, 734-745. doi: 10.1016/j.molcel.2016.09.038.
- Abstract
- The eukaryotic RNA exosome is an essential and conserved 3'-to-5' exoribonuclease complex that degrades or processes nearly every class of cellular RNA. The nuclear RNA exosome includes a 9-subunit non-catalytic core that binds Rrp44 (Dis3) and Rrp6 subunits to modulate their processive and distributive 3'-to-5' exoribonuclease activities, respectively. Here we utilize an engineered RNA with two 3' ends to obtain a crystal structure of an 11-subunit nuclear exosome bound to RNA at 3.1 Å. The structure reveals an extended RNA path to Rrp6 that penetrates into the non-catalytic core; contacts between the non-catalytic core and Rrp44, which inhibit exoribonuclease activity; and features of the Rrp44 exoribonuclease site that support its ability to degrade 3' phosphate RNA substrates. Using reconstituted exosome complexes, we show that 3' phosphate RNA is not a substrate for Rrp6 but is readily degraded by Rrp44 in the nuclear exosome.