Summary information and primary citation
- PDB-id
- 5keg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.2 Å)
- Summary
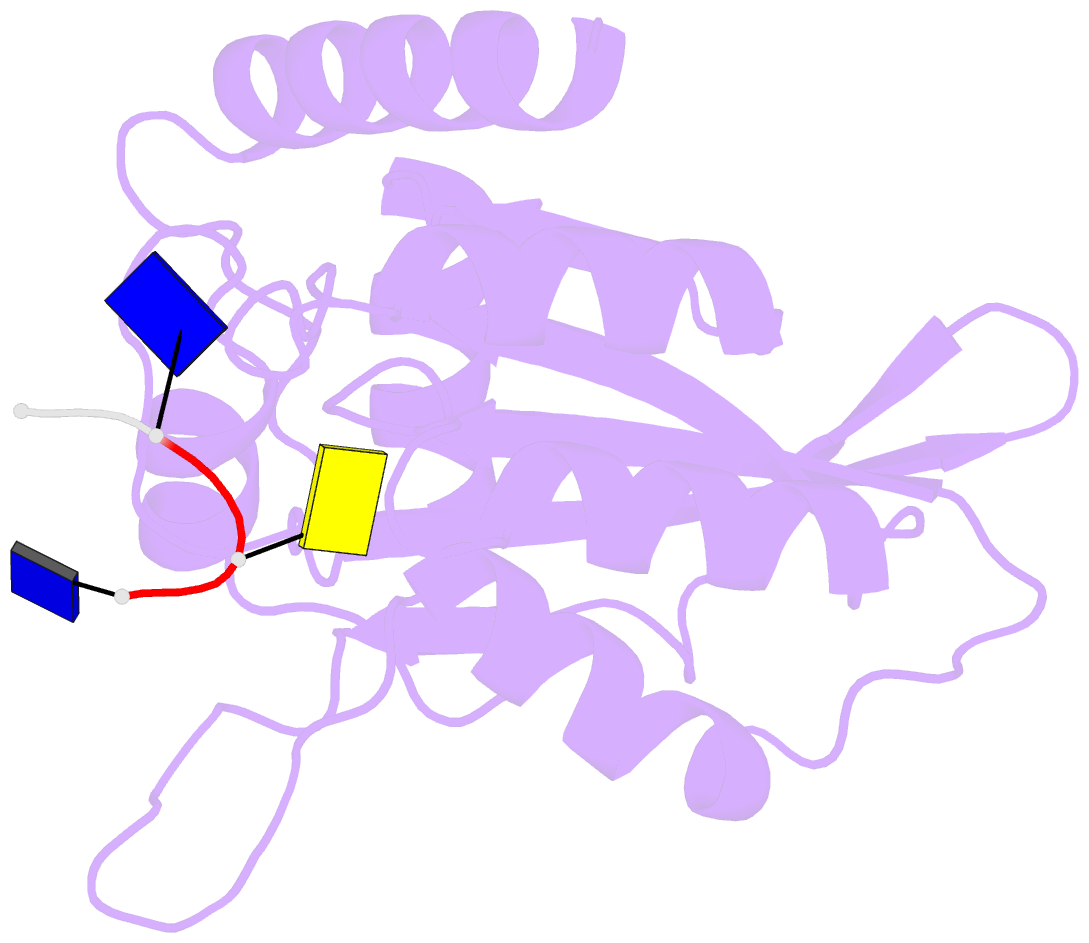
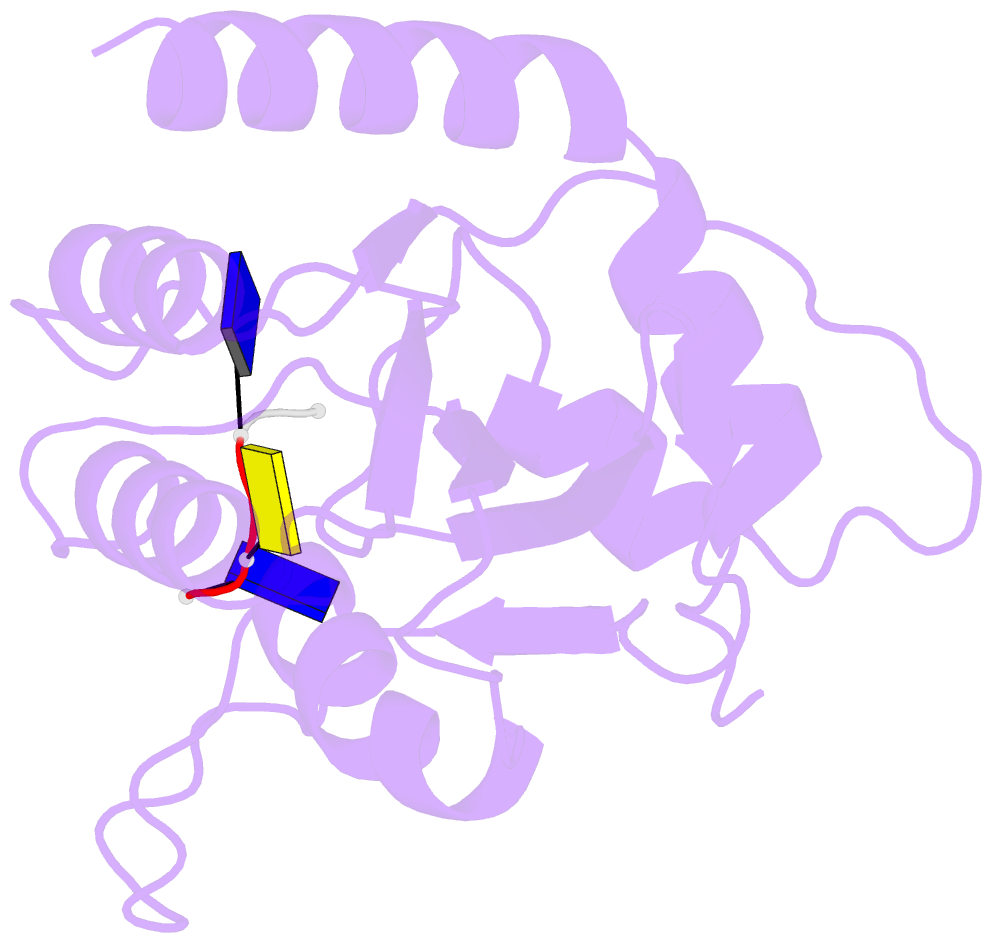
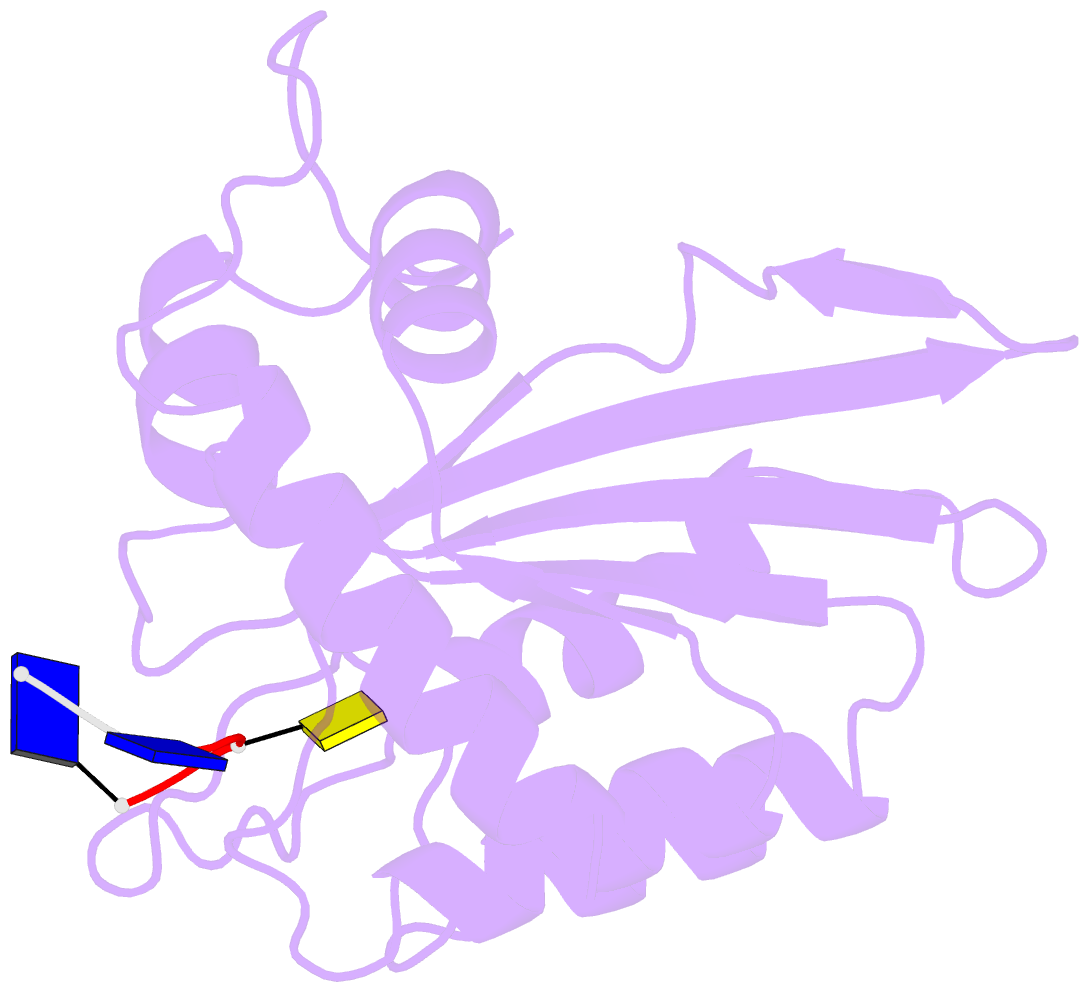
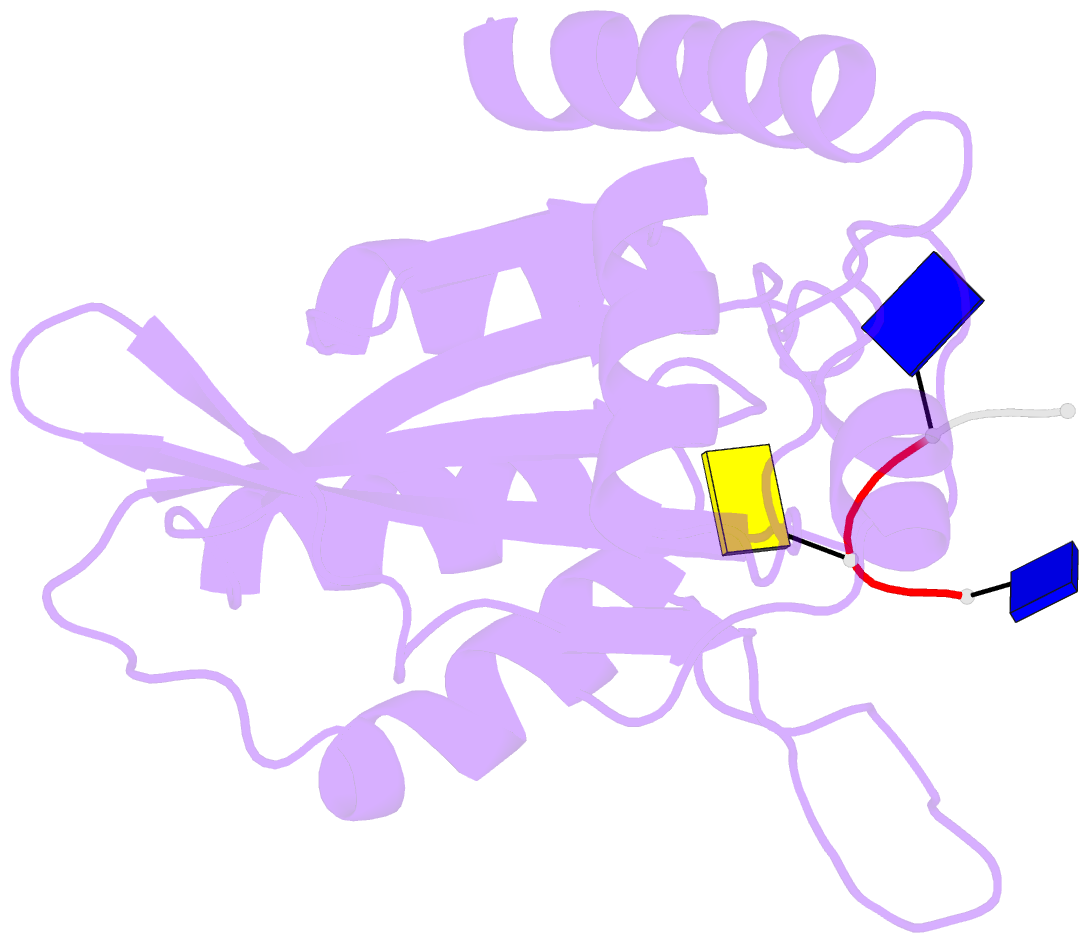
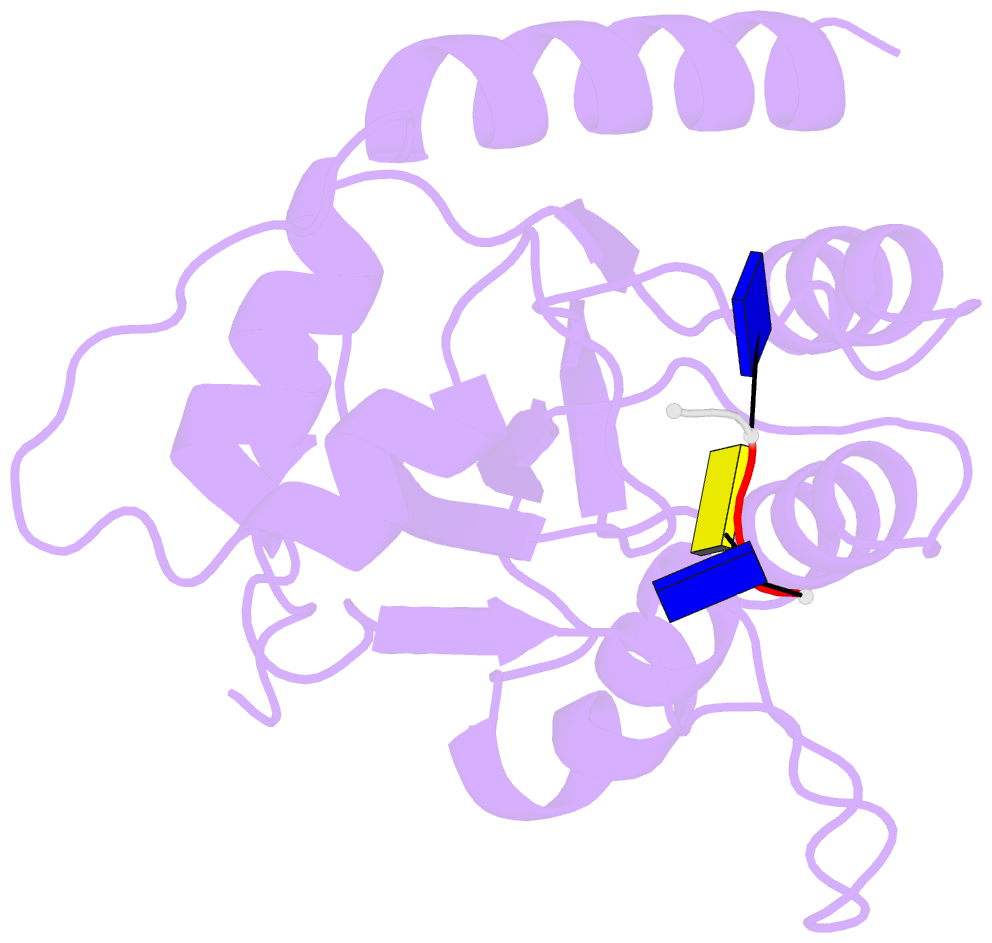
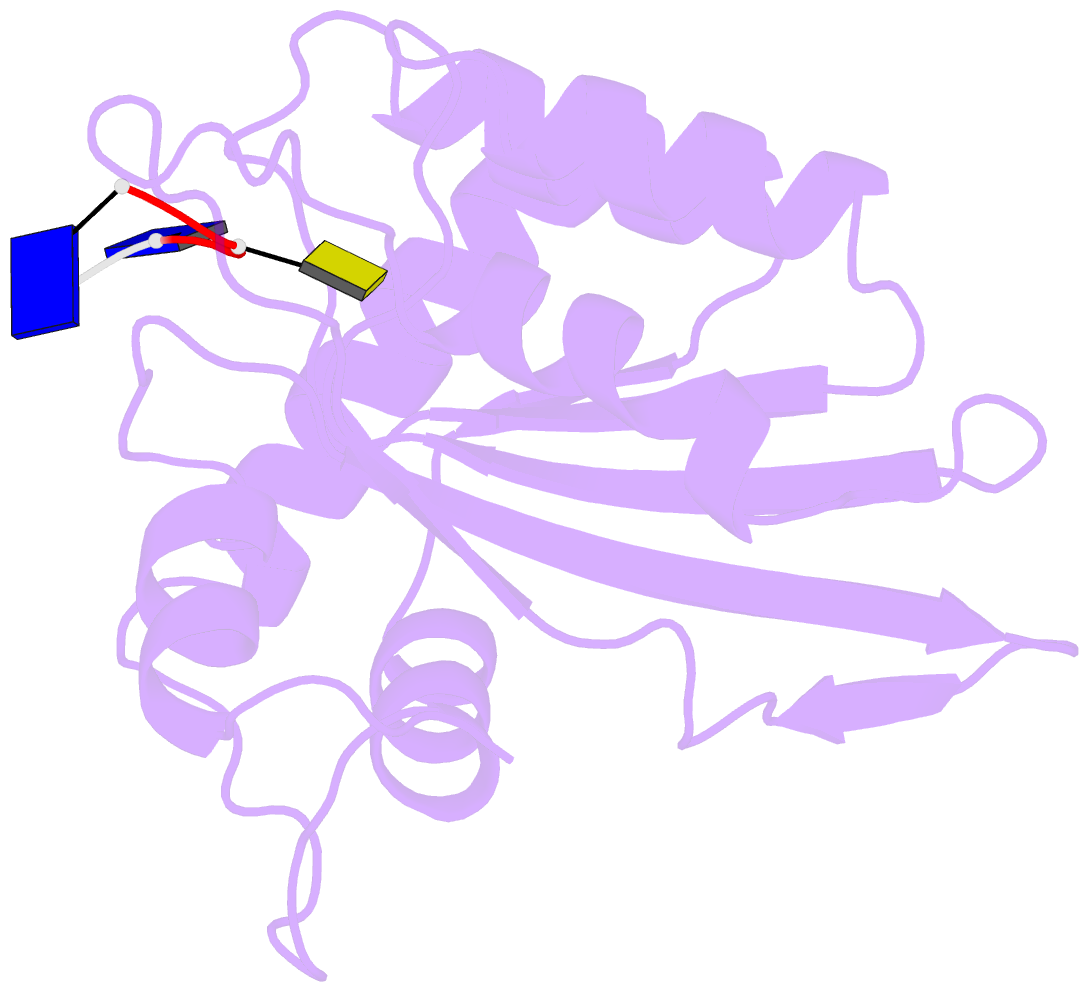
- Crystal structure of apobec3a in complex with a single-stranded DNA
- Reference
- Kouno T, Silvas TV, Hilbert BJ, Shandilya SMD, Bohn MF, Kelch BA, Royer WE, Somasundaran M, Kurt Yilmaz N, Matsuo H, Schiffer CA (2017): "Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity." Nat Commun, 8, 15024. doi: 10.1038/ncomms15024.
- Abstract
- Nucleic acid editing enzymes are essential components of the immune system that lethally mutate viral pathogens and somatically mutate immunoglobulins, and contribute to the diversification and lethality of cancers. Among these enzymes are the seven human APOBEC3 deoxycytidine deaminases, each with unique target sequence specificity and subcellular localization. While the enzymology and biological consequences have been extensively studied, the mechanism by which APOBEC3s recognize and edit DNA remains elusive. Here we present the crystal structure of a complex of a cytidine deaminase with ssDNA bound in the active site at 2.2 Å. This structure not only visualizes the active site poised for catalysis of APOBEC3A, but pinpoints the residues that confer specificity towards CC/TC motifs. The APOBEC3A-ssDNA complex defines the 5'-3' directionality and subtle conformational changes that clench the ssDNA within the binding groove, revealing the architecture and mechanism of ssDNA recognition that is likely conserved among all polynucleotide deaminases, thereby opening the door for the design of mechanistic-based therapeutics.