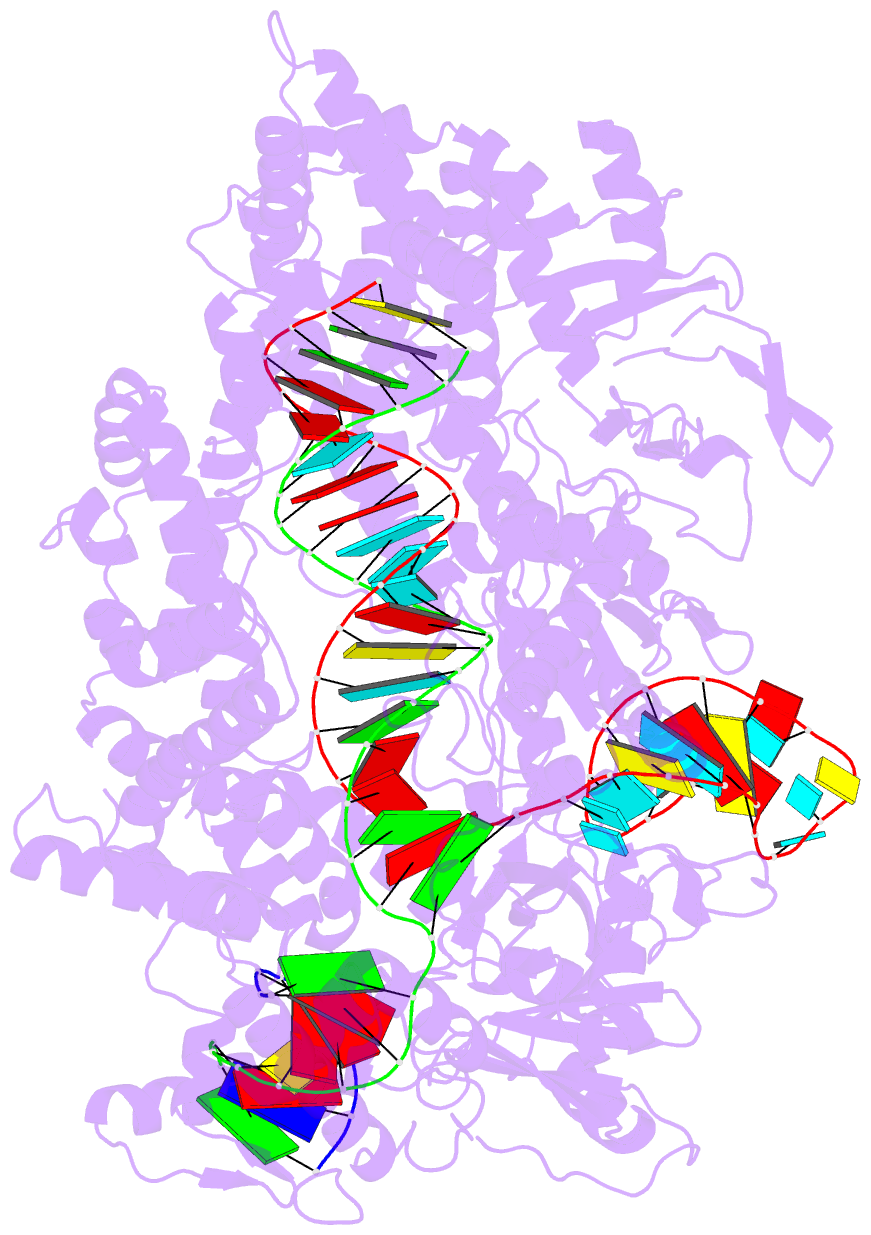
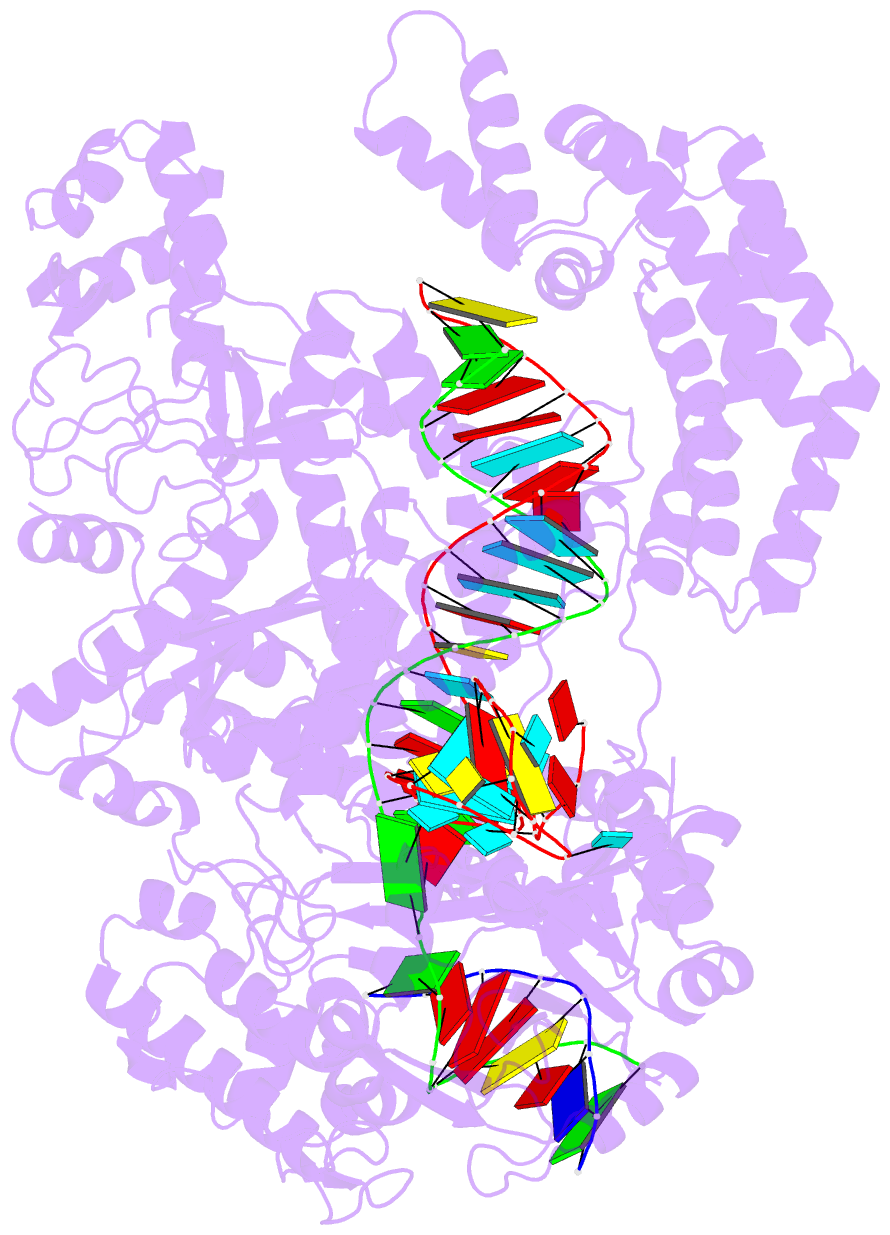
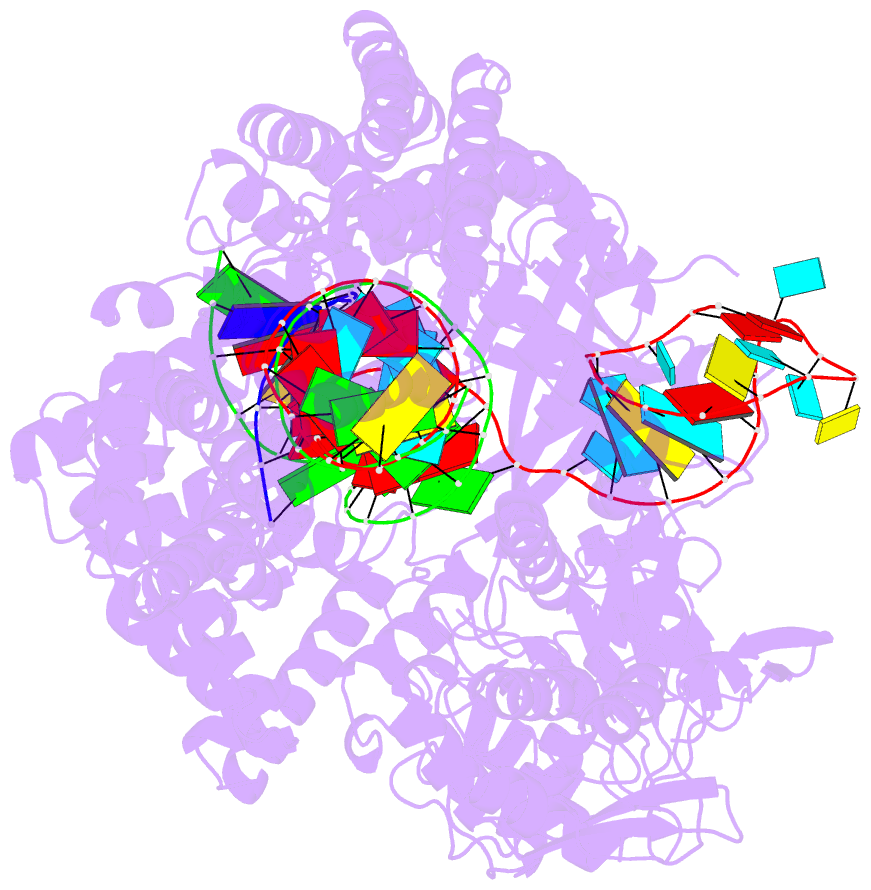
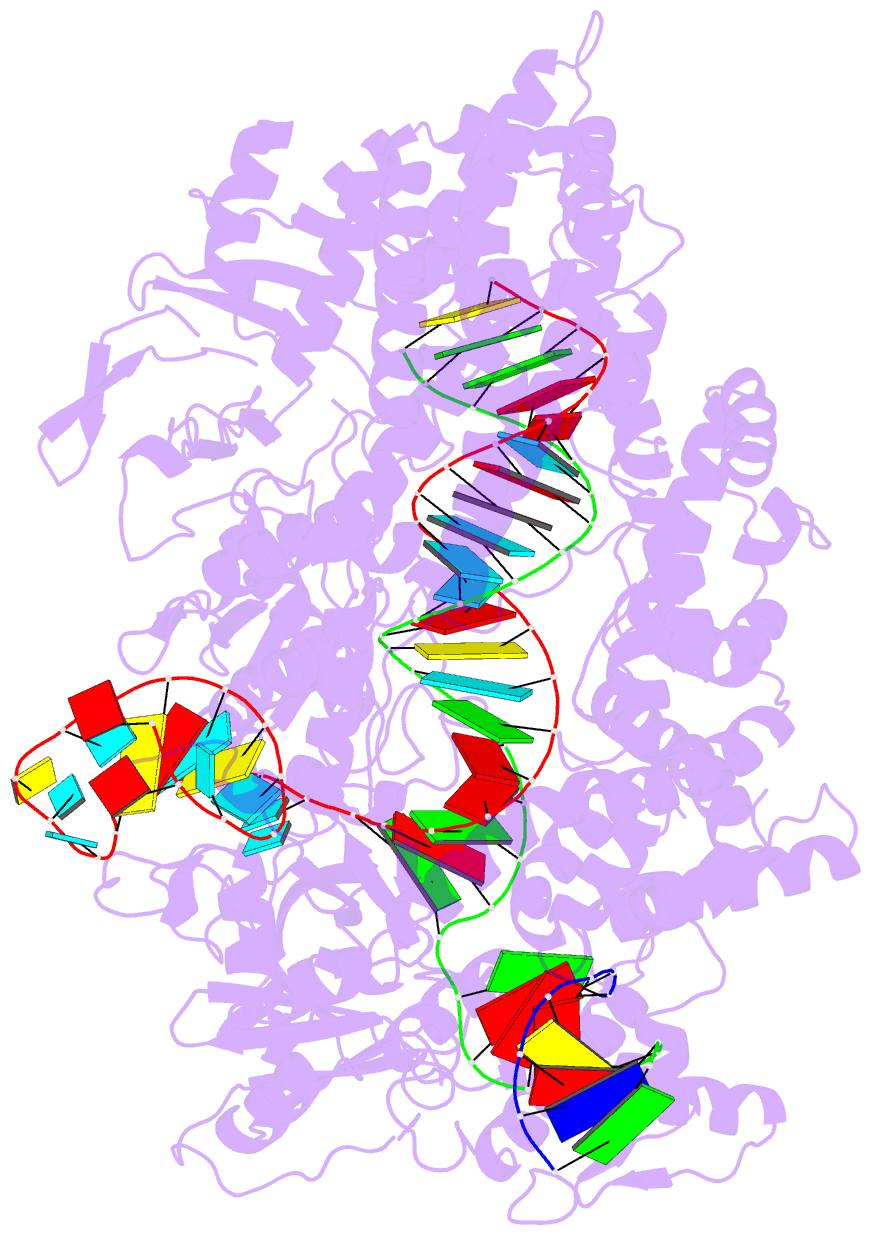
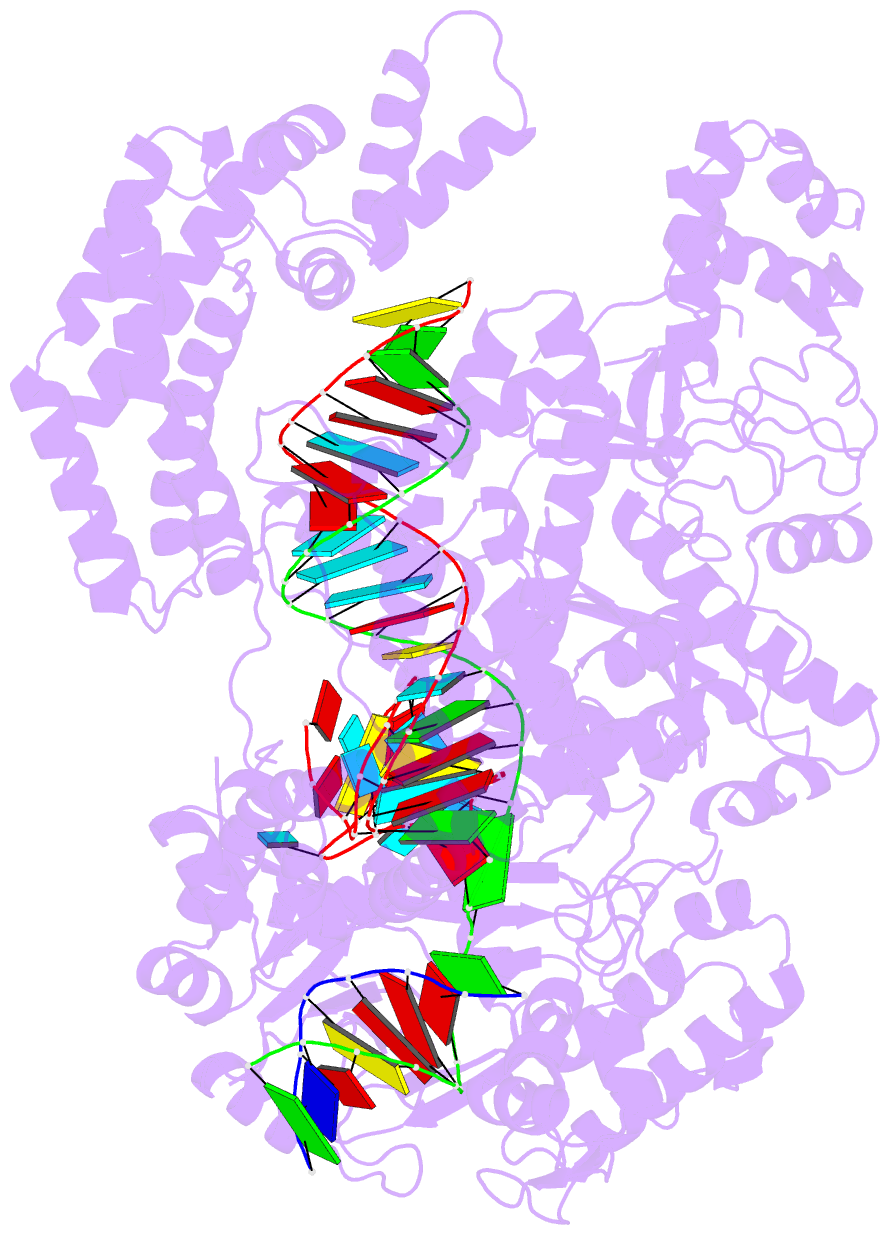
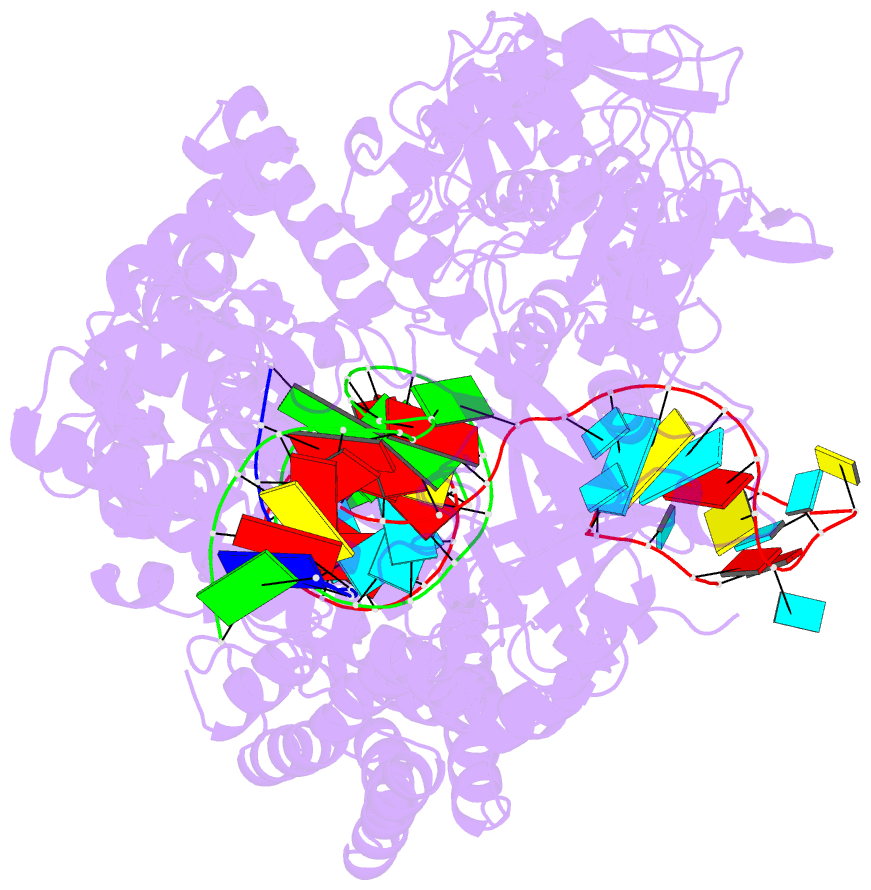
Summary information and primary citation
- PDB-id
- 5kk5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA-RNA
- Method
- X-ray (3.289 Å)
- Summary
- Ascpf1(e993a)-crrna-DNA ternary complex
- Reference
- Gao P, Yang H, Rajashankar KR, Huang Z, Patel DJ (2016): "Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition." Cell Res., 26, 901-913. doi: 10.1038/cr.2016.88.
- Abstract
- CRISPR-Cas9 and CRISPR-Cpf1 systems have been successfully harnessed for genome editing. In the CRISPR-Cas9 system, the preordered A-form RNA seed sequence and preformed protein PAM-interacting cleft are essential for Cas9 to form a DNA recognition-competent structure. Whether the CRISPR-Cpf1 system employs a similar mechanism for target DNA recognition remains unclear. Here, we have determined the crystal structure of Acidaminococcus sp. Cpf1 (AsCpf1) in complex with crRNA and target DNA. Structural comparison between the AsCpf1-crRNA-DNA ternary complex and the recently reported Lachnospiraceae bacterium Cpf1 (LbCpf1)-crRNA binary complex identifies a unique mechanism employed by Cpf1 for target recognition. The seed sequence required for initial DNA interrogation is disordered in the Cpf1-cRNA binary complex, but becomes ordered upon ternary complex formation. Further, the PAM interacting cleft of Cpf1 undergoes an "open-to-closed" conformational change upon target DNA binding, which in turn induces structural changes within Cpf1 to accommodate the ordered A-form seed RNA segment. This unique mechanism of target recognition by Cpf1 is distinct from that reported previously for Cas9.