Summary information and primary citation
- PDB-id
- 5kl8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-binding protein-RNA
- Method
- X-ray (4.0 Å)
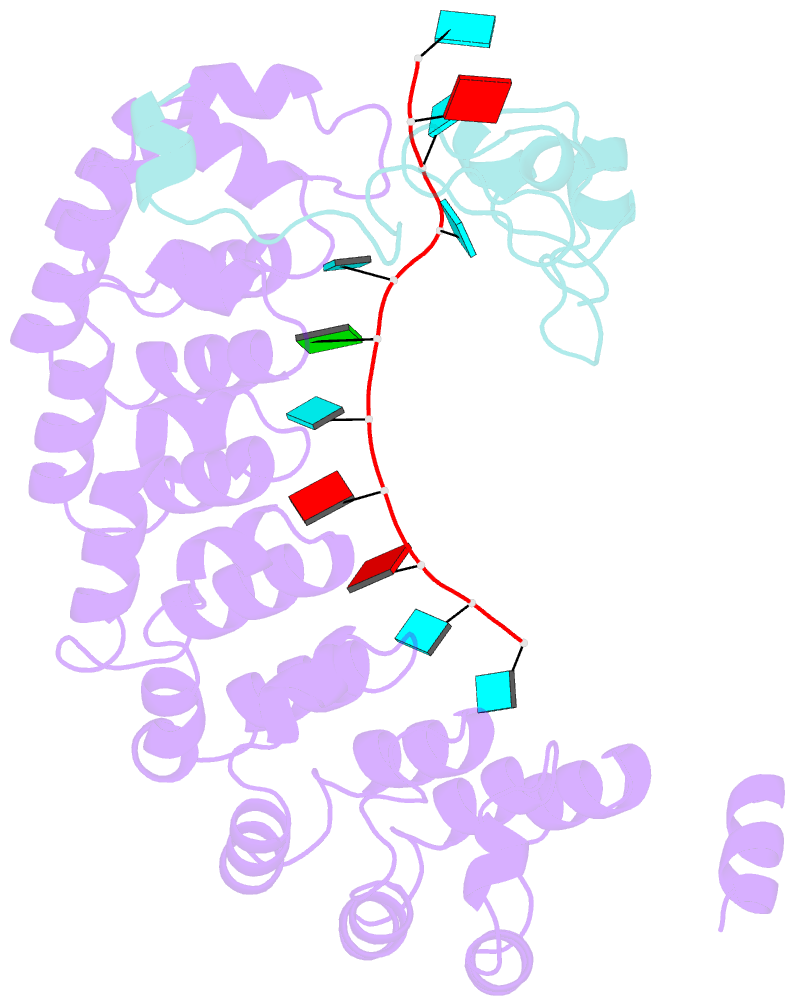
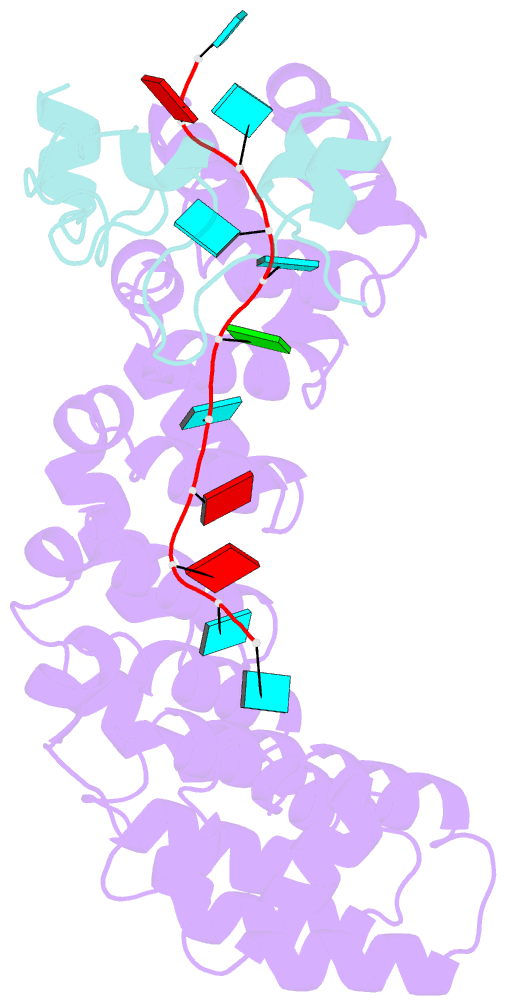
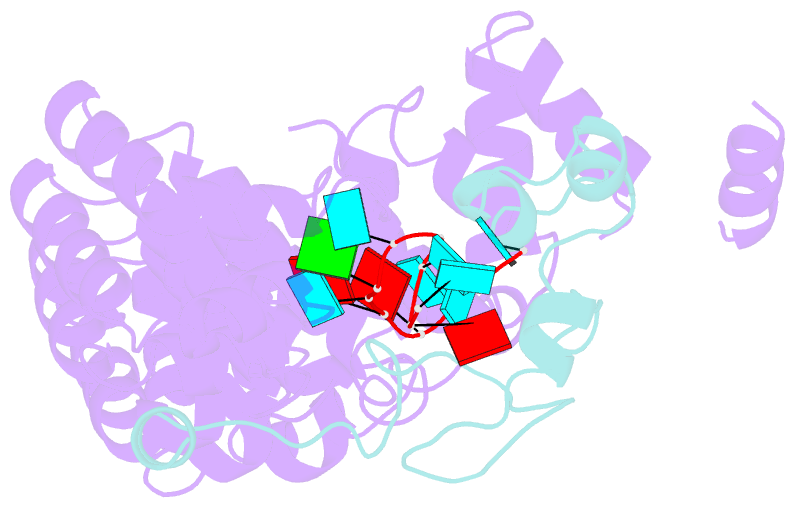
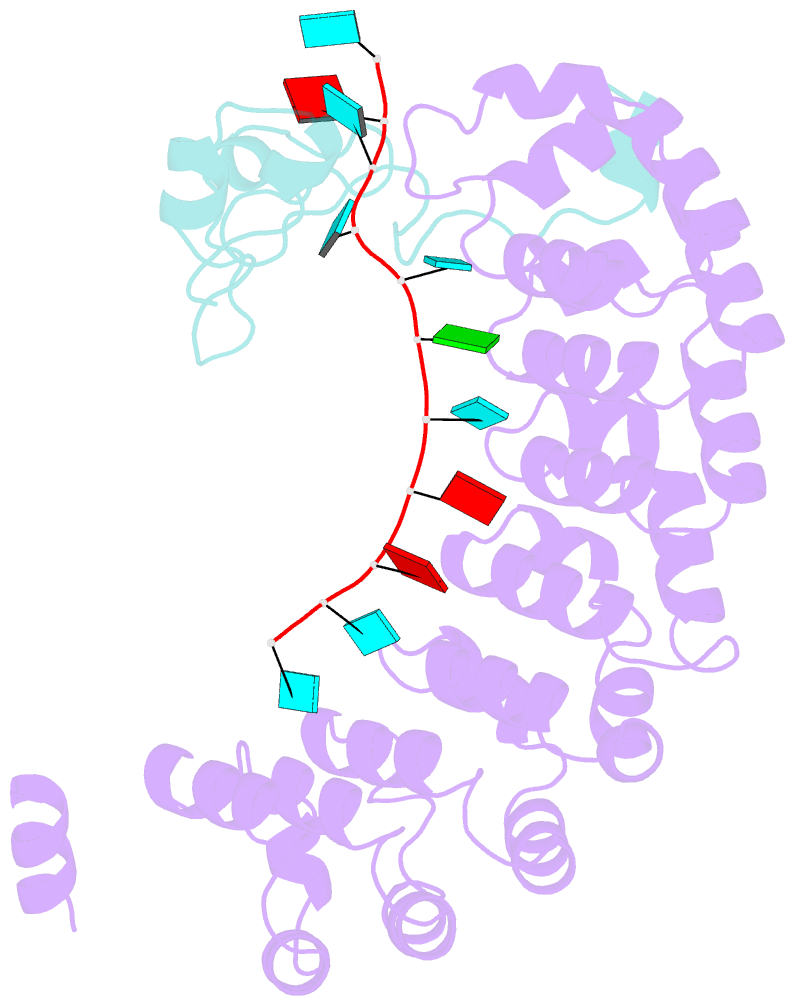
- Summary
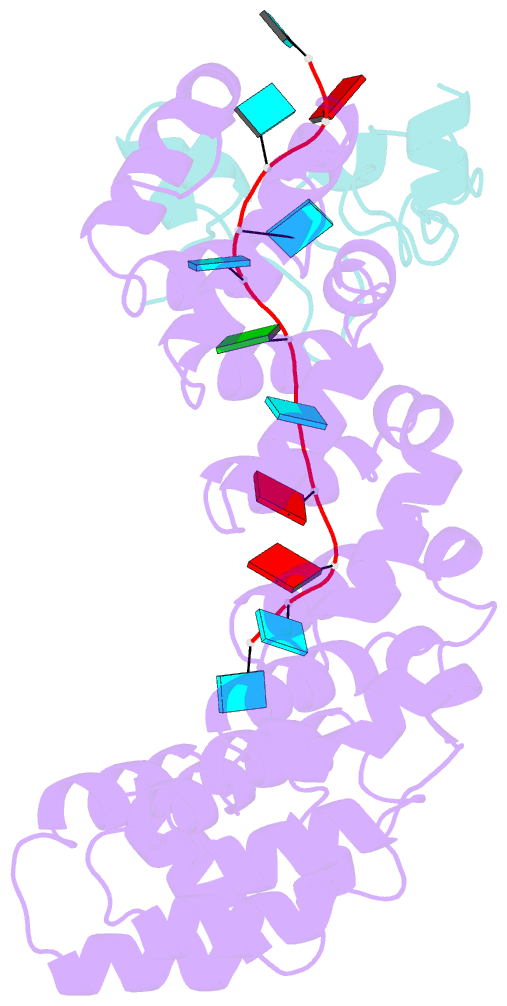
- Crystal structure of the pumilio-nos-cyclinb RNA complex
- Reference
- Weidmann CA, Qiu C, Arvola RM, Lou TF, Killingsworth J, Campbell ZT, Tanaka Hall TM, Goldstrohm AC (2016): "Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio." Elife, 5. doi: 10.7554/eLife.17096.
- Abstract
- Collaboration among the multitude of RNA-binding proteins (RBPs) is ubiquitous, yet our understanding of these key regulatory complexes has been limited to single RBPs. We investigated combinatorial translational regulation by Drosophila Pumilio (Pum) and Nanos (Nos), which control development, fertility, and neuronal functions. Our results show how the specificity of one RBP (Pum) is modulated by cooperative RNA recognition with a second RBP (Nos) to synergistically repress mRNAs. Crystal structures of Nos-Pum-RNA complexes reveal that Nos embraces Pum and RNA, contributes sequence-specific contacts, and increases Pum RNA-binding affinity. Nos shifts the recognition sequence and promotes repression complex formation on mRNAs that are not stably bound by Pum alone, explaining the preponderance of sub-optimal Pum sites regulated in vivo. Our results illuminate the molecular mechanism of a regulatory switch controlling crucial gene expression programs, and provide a framework for understanding how the partnering of RBPs evokes changes in binding specificity that underlie regulatory network dynamics.