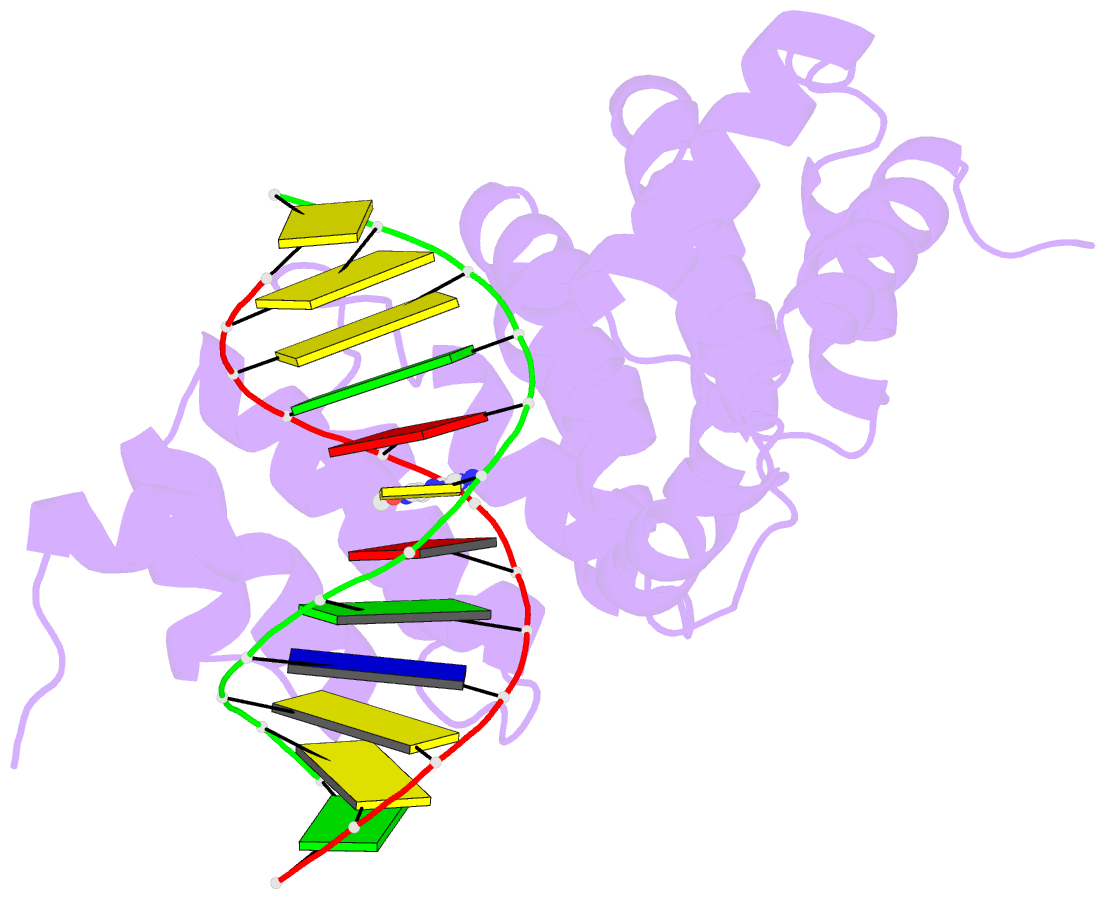
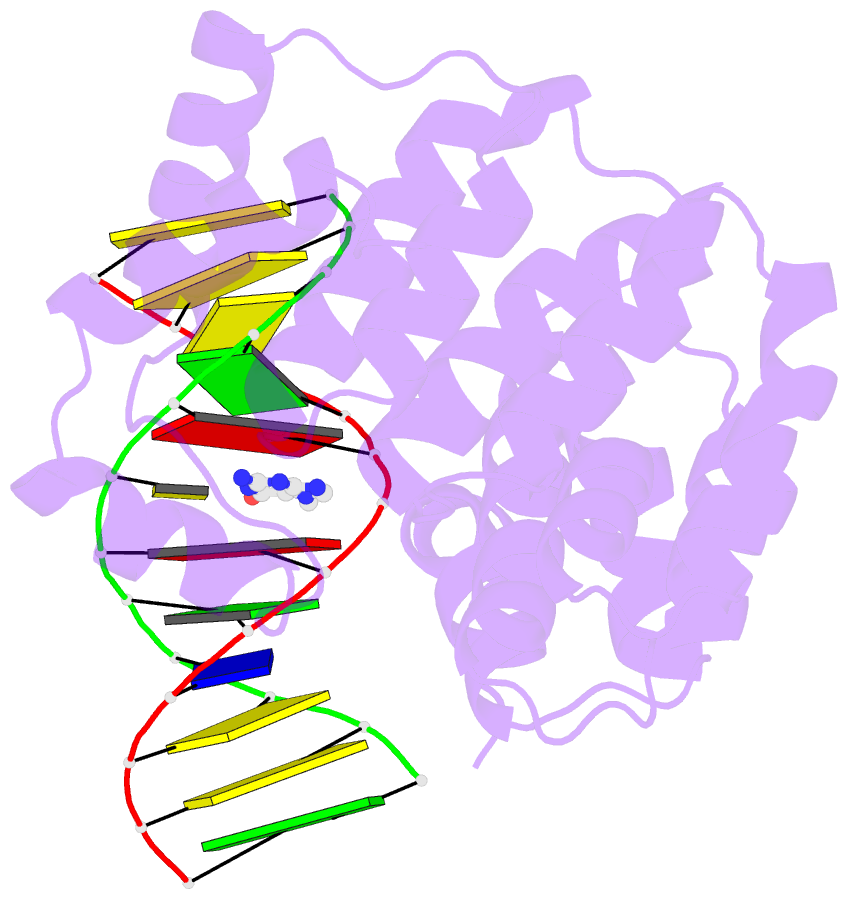
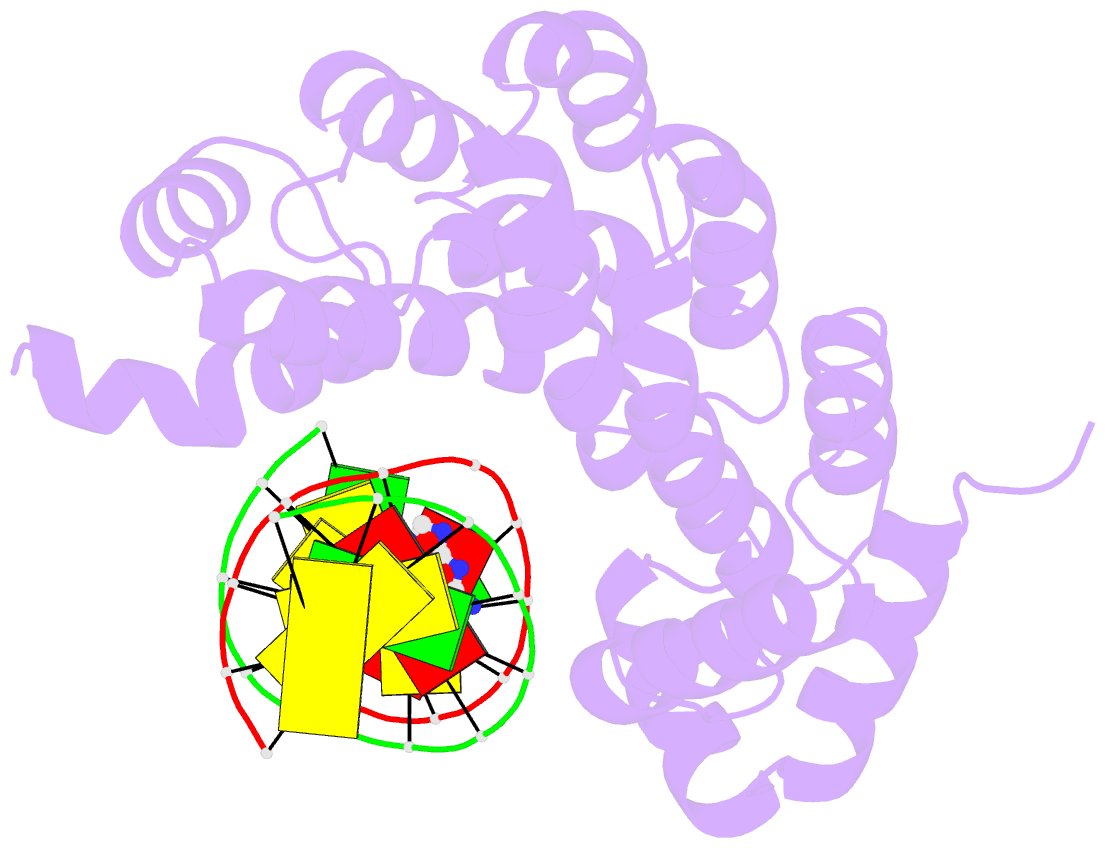
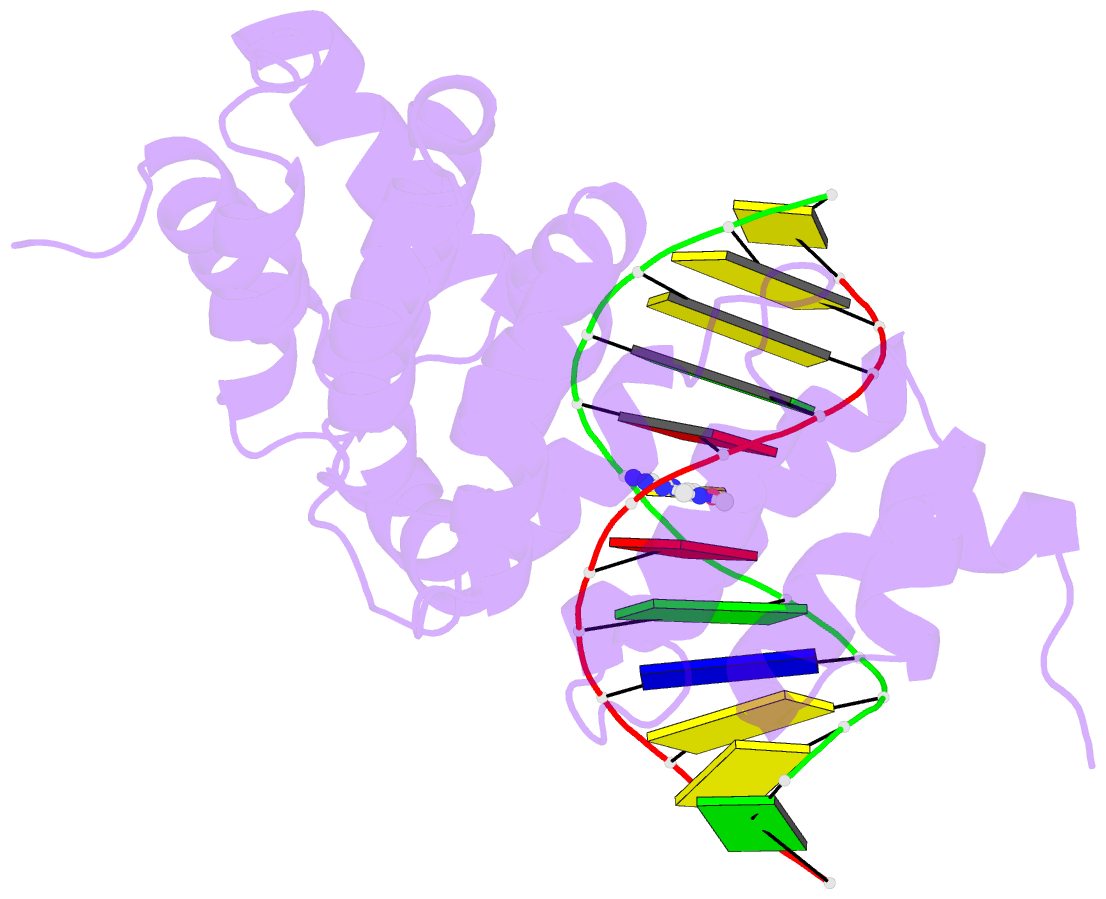
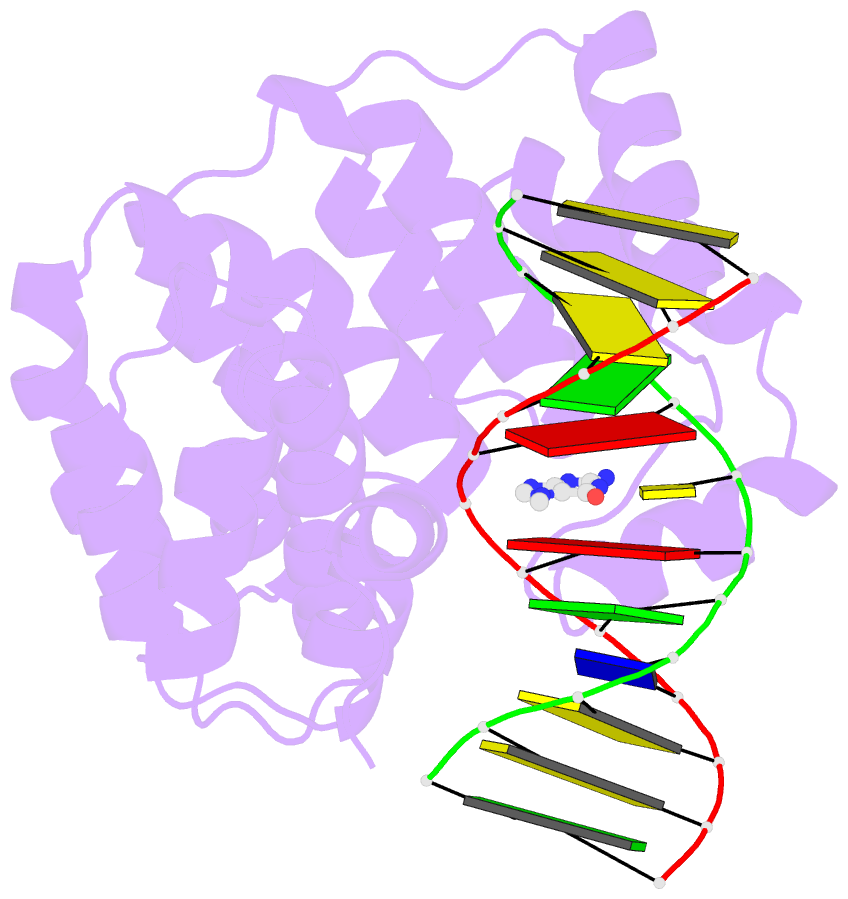
Summary information and primary citation
- PDB-id
- 5kub; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.73 Å)
- Summary
- Bacillus cereus DNA glycosylase alkd bound to 7-methylguanine nucleobase and DNA containing an oxocarbenium-intermediate analog
- Reference
- Parsons ZD, Bland JM, Mullins EA, Eichman BF (2016): "A Catalytic Role for C-H/ pi Interactions in Base Excision Repair by Bacillus cereus DNA Glycosylase AlkD." J.Am.Chem.Soc., 138, 11485-11488. doi: 10.1021/jacs.6b07399.
- Abstract
- DNA glycosylases protect genomic integrity by locating and excising aberrant nucleobases. Substrate recognition and excision usually take place in an extrahelical conformation, which is often stabilized by π-stacking interactions between the lesion nucleobase and aromatic side chains in the glycosylase active site. Bacillus cereus AlkD is the only DNA glycosylase known to catalyze base excision without extruding the damaged nucleotide from the DNA helix. Instead of contacting the nucleobase itself, the AlkD active site interacts with the lesion deoxyribose through a series of C-H/π interactions. These interactions are ubiquitous in protein structures, but evidence for their catalytic significance in enzymology is lacking. Here, we show that the C-H/π interactions between AlkD and the lesion deoxyribose participate in catalysis of glycosidic bond cleavage. This is the first demonstration of a catalytic role for C-H/π interactions as intermolecular forces important to DNA repair.