Summary information and primary citation
- PDB-id
- 5ld2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- cryo-EM (3.83 Å)
- Summary
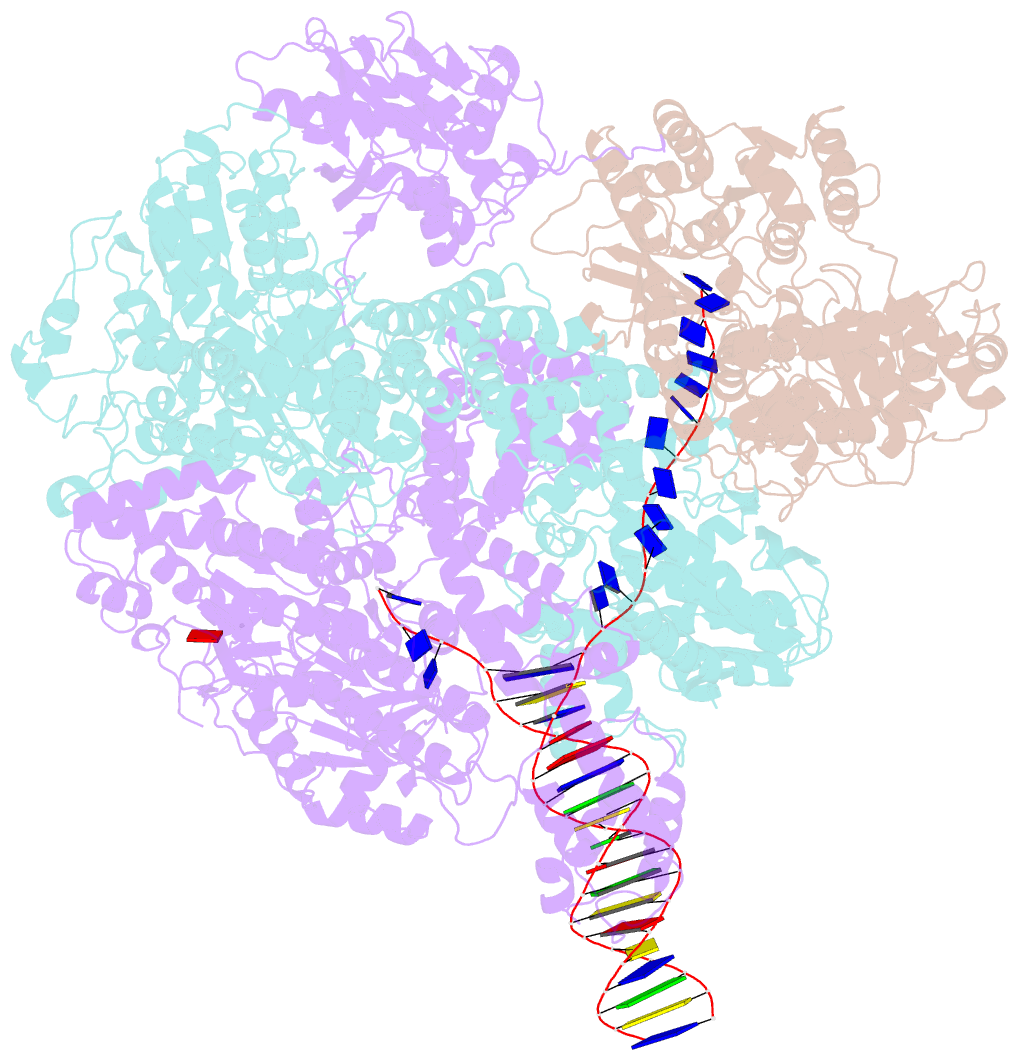
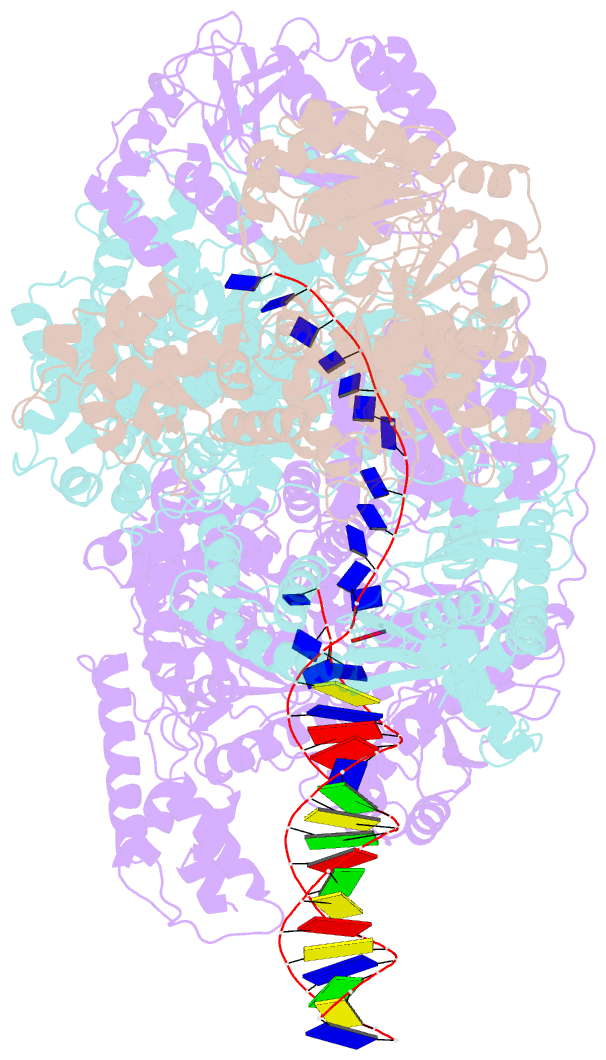
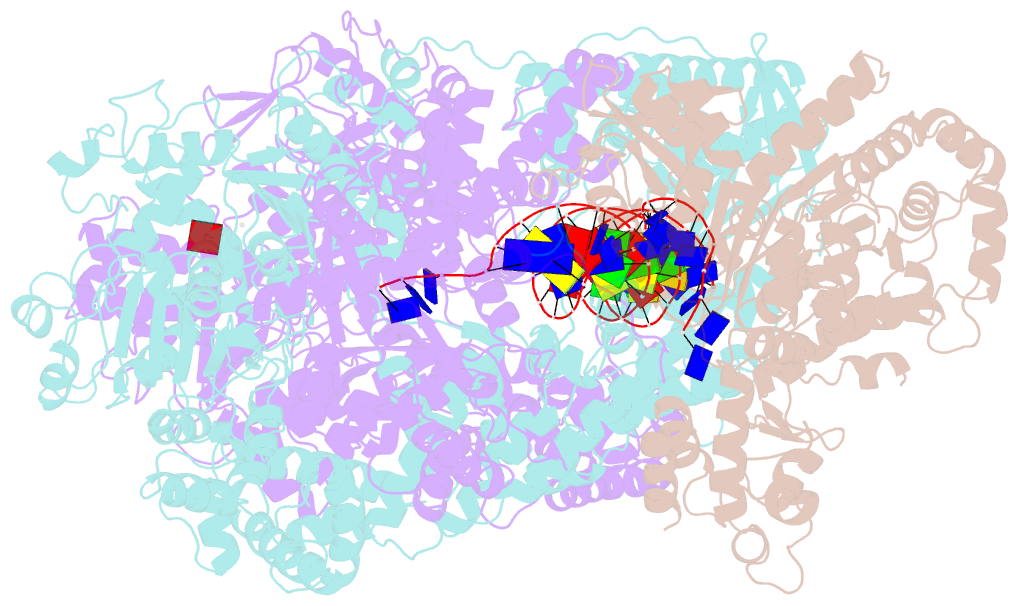
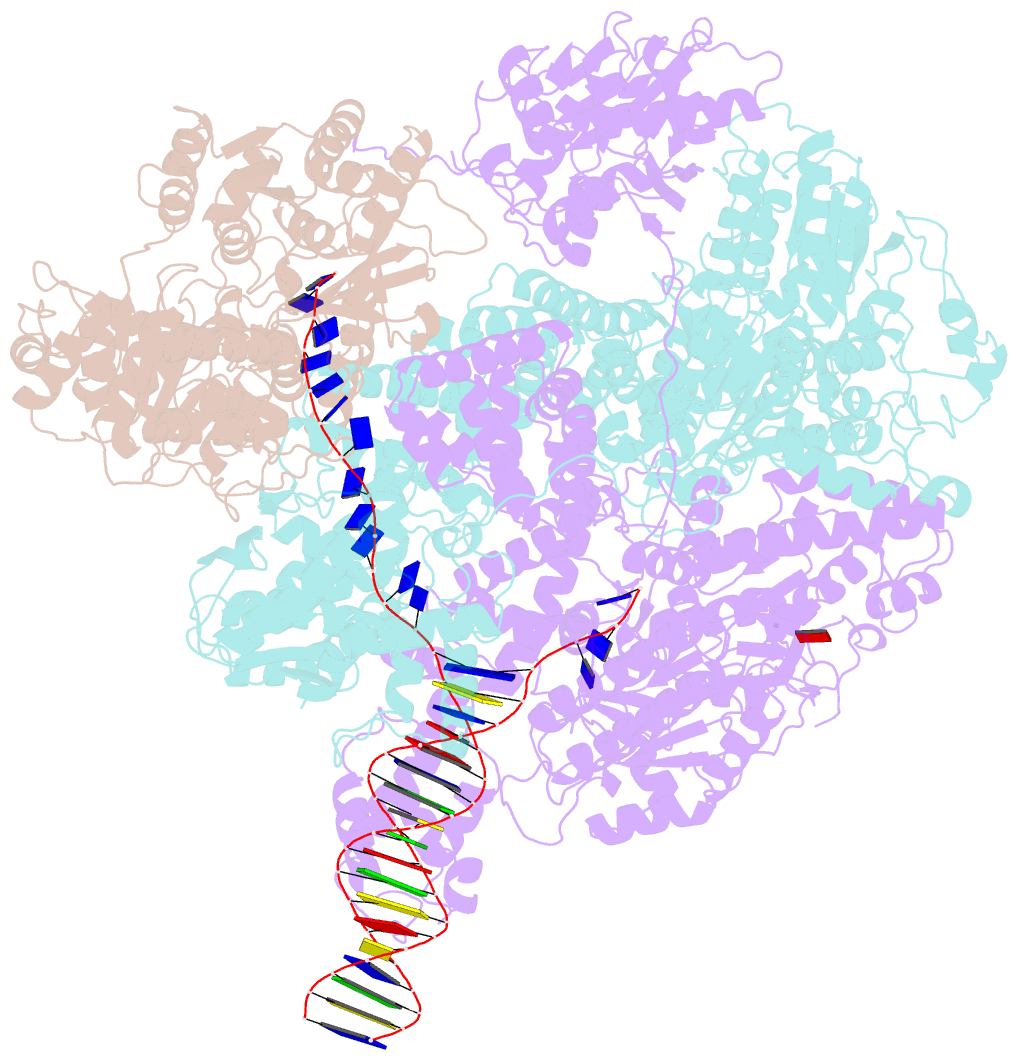
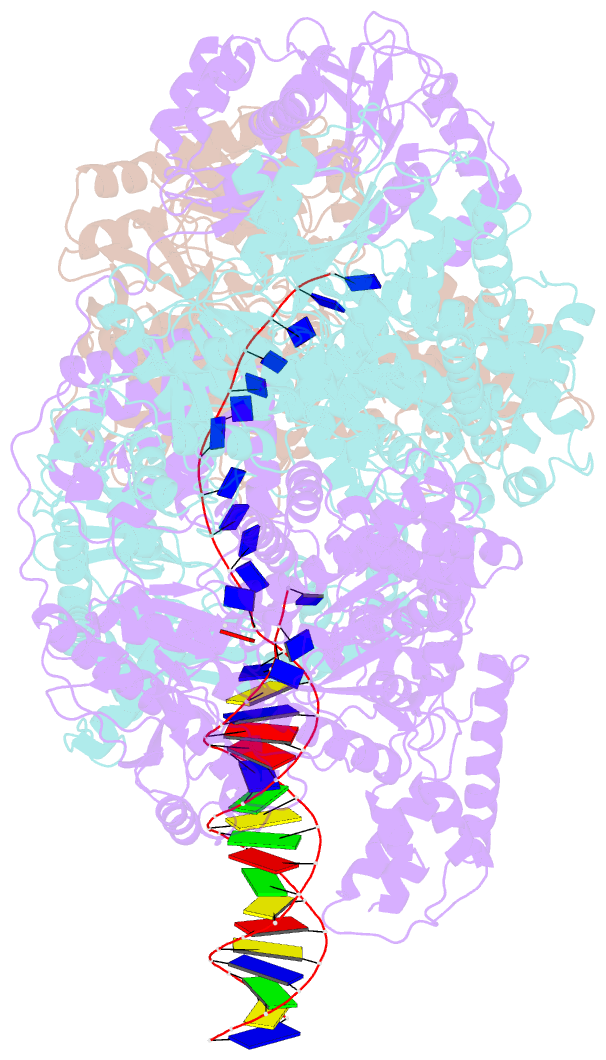
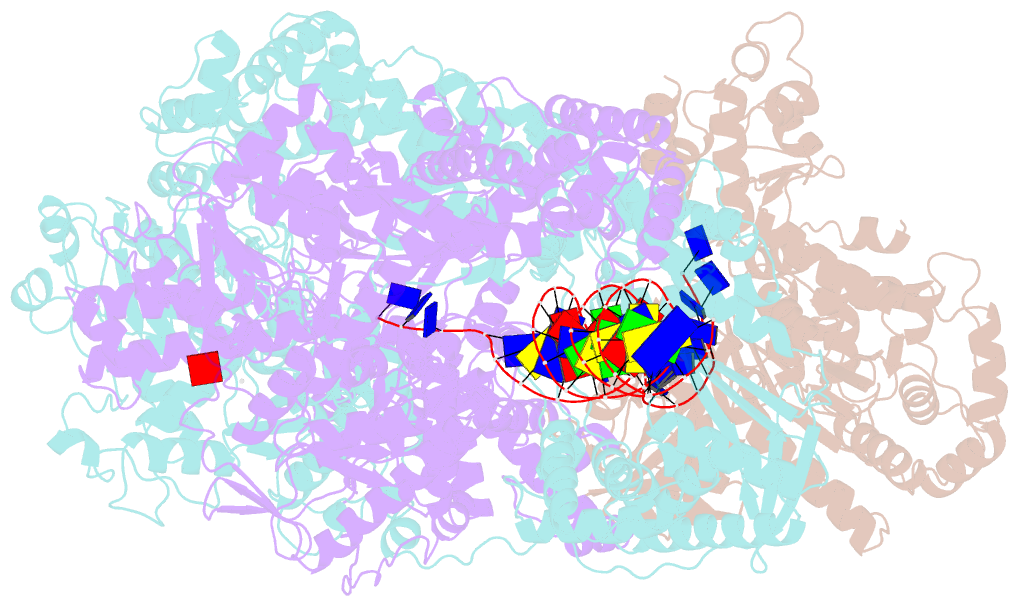
- cryo-EM structure of recbcd+DNA complex revealing activated nuclease domain
- Reference
- Wilkinson M, Chaban Y, Wigley DB (2016): "Mechanism for nuclease regulation in RecBCD." Elife, 5. doi: 10.7554/eLife.18227.
- Abstract
- In bacterial cells, processing of double-stranded DNA breaks for repair by homologous recombination is catalysed by AddAB, AdnAB or RecBCD-type helicase-nucleases. These enzyme complexes are highly processive, duplex unwinding and degrading machines that require tight regulation. Here, we report the structure of E.coli RecBCD, determined by cryoEM at 3.8 Å resolution, with a DNA substrate that reveals how the nuclease activity of the complex is activated once unwinding progresses. Extension of the 5'-tail of the unwound duplex induces a large conformational change in the RecD subunit, that is transferred through the RecC subunit to activate the nuclease domain of the RecB subunit. The process involves a SH3 domain that binds to a region of the RecB subunit in a binding mode that is distinct from others observed previously in SH3 domains and, to our knowledge, this is the first example of peptide-binding of an SH3 domain in a bacterial system.