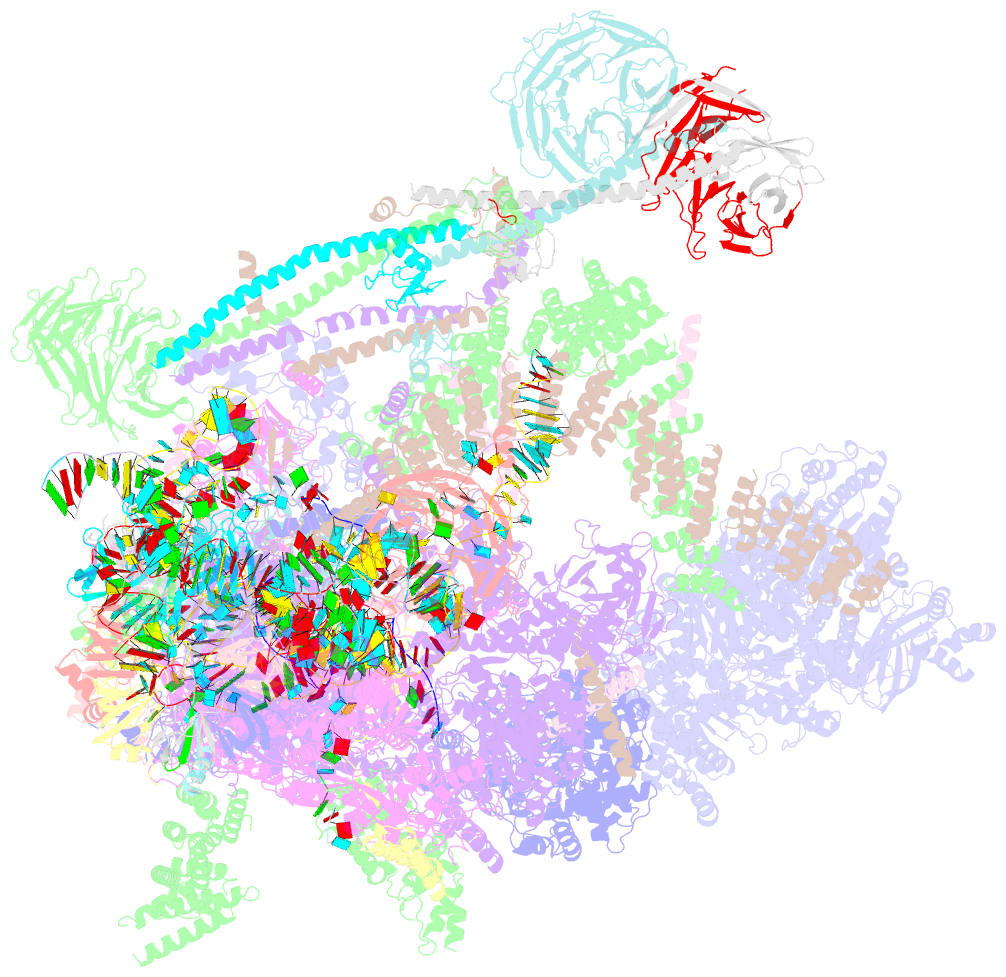
Summary information and primary citation
- PDB-id
- 5lj5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- cryo-EM (10.0 Å)
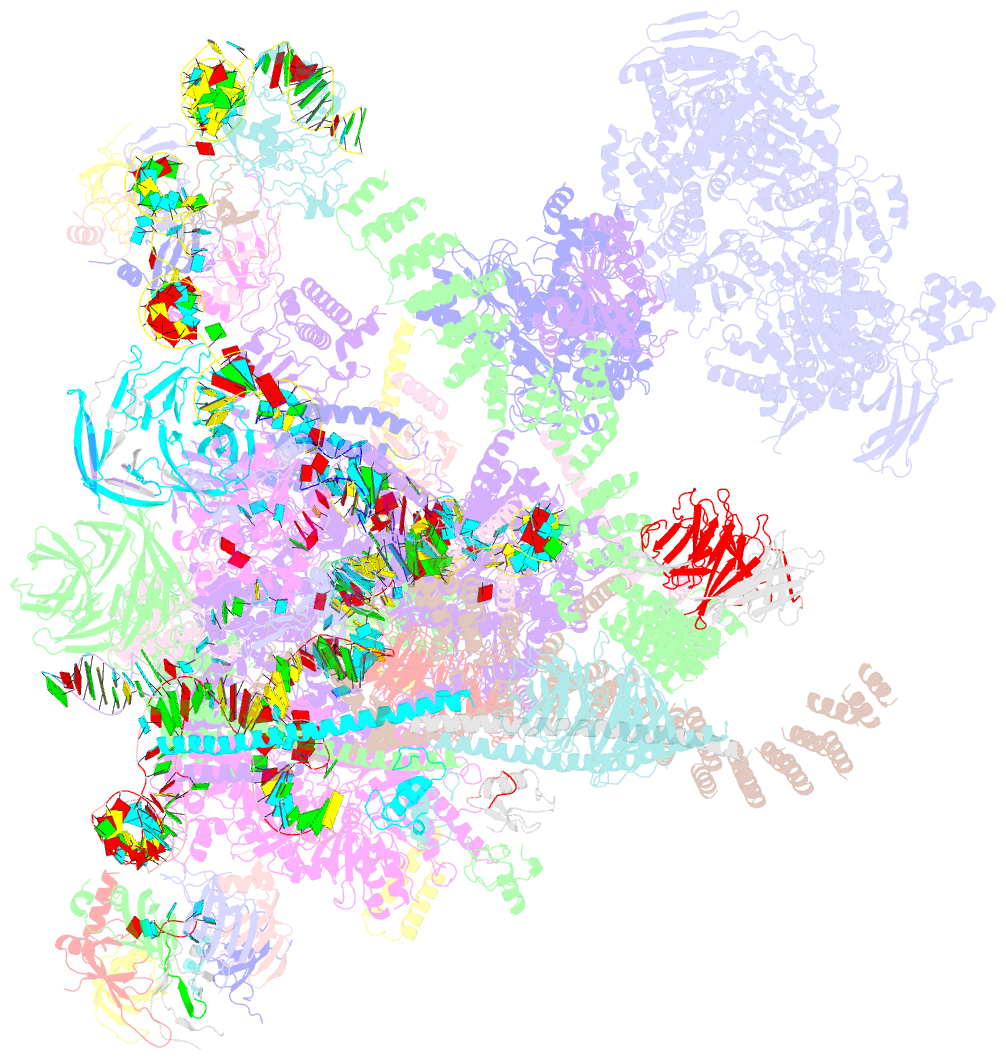
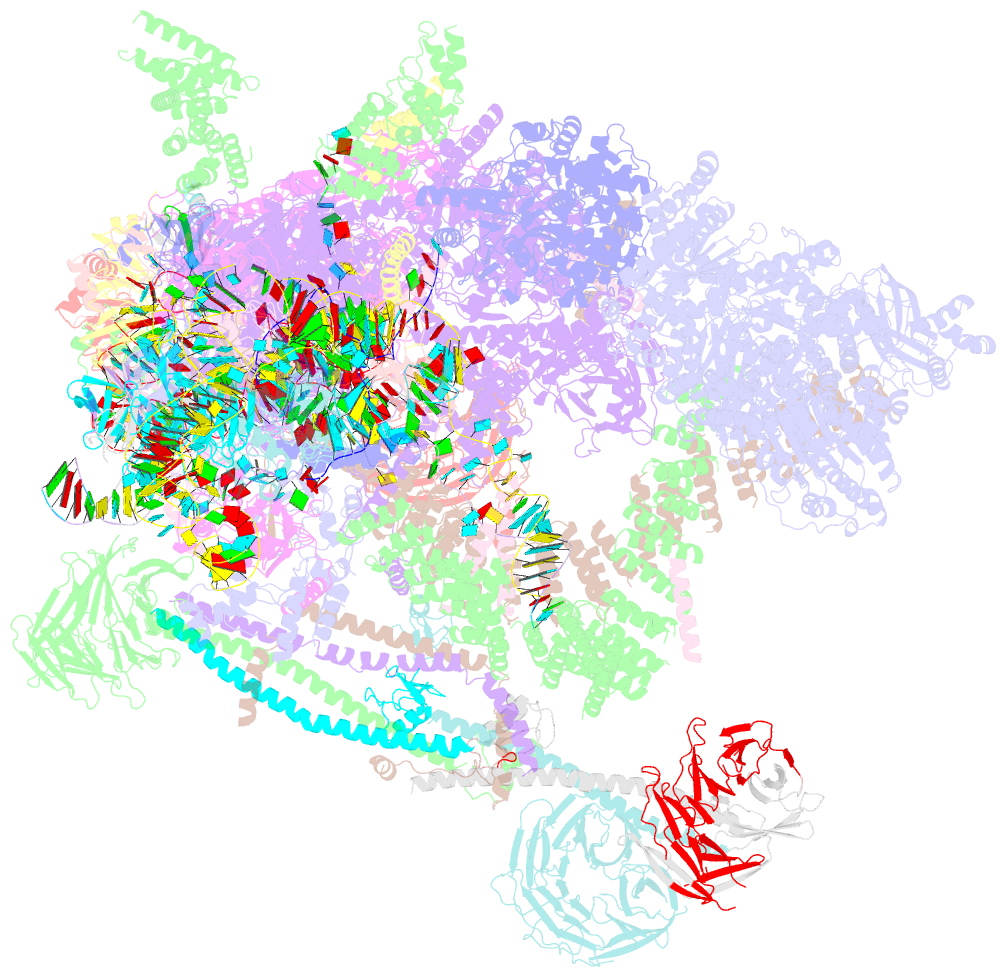
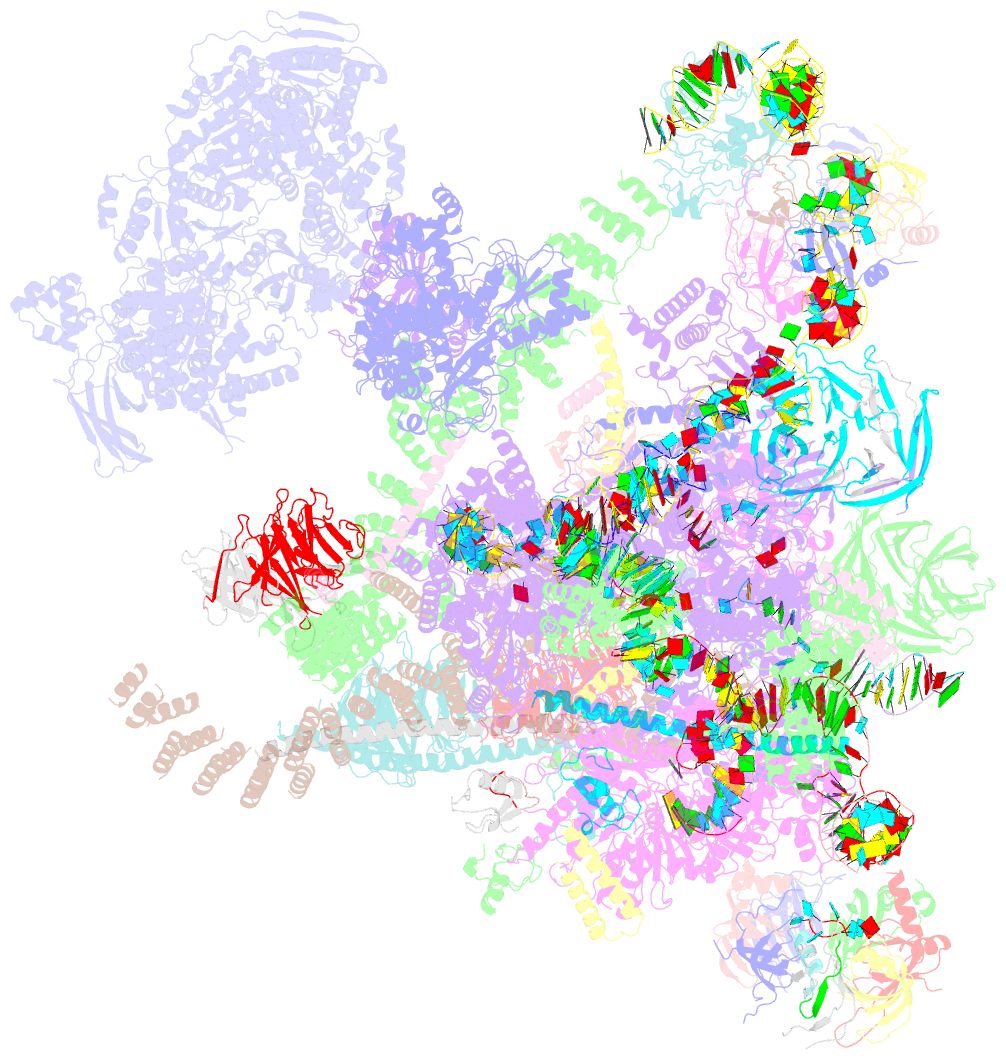
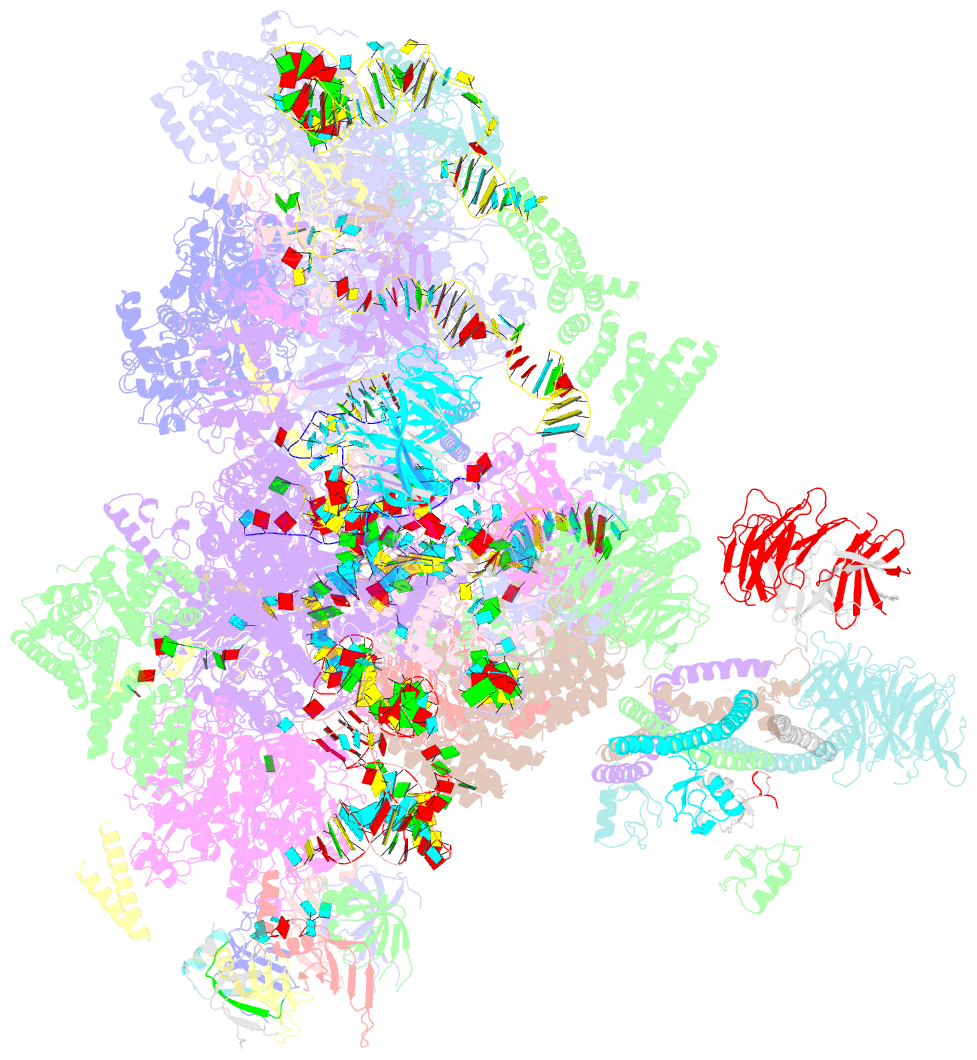
- Summary
- Overall structure of the yeast spliceosome immediately after branching.
- Reference
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K (2016): "Cryo-EM structure of the spliceosome immediately after branching." Nature, 537, 197-201. doi: 10.1038/nature19316.
- Abstract
- Precursor mRNA (pre-mRNA) splicing proceeds by two consecutive transesterification reactions via a lariat-intron intermediate. Here we present the 3.8 Å cryo-electron microscopy structure of the spliceosome immediately after lariat formation. The 5'-splice site is cleaved but remains close to the catalytic Mg2+ site in the U2/U6 small nuclear RNA (snRNA) triplex, and the 5'-phosphate of the intron nucleotide G(+1) is linked to the branch adenosine 2'OH. The 5'-exon is held between the Prp8 amino-terminal and linker domains, and base-pairs with U5 snRNA loop 1. Non-Watson-Crick interactions between the branch helix and 5'-splice site dock the branch adenosine into the active site, while intron nucleotides +3 to +6 base-pair with the U6 snRNA ACAGAGA sequence. Isy1 and the step-one factors Yju2 and Cwc25 stabilize docking of the branch helix. The intron downstream of the branch site emerges between the Prp8 reverse transcriptase and linker domains and extends towards the Prp16 helicase, suggesting a plausible mechanism of remodelling before exon ligation.