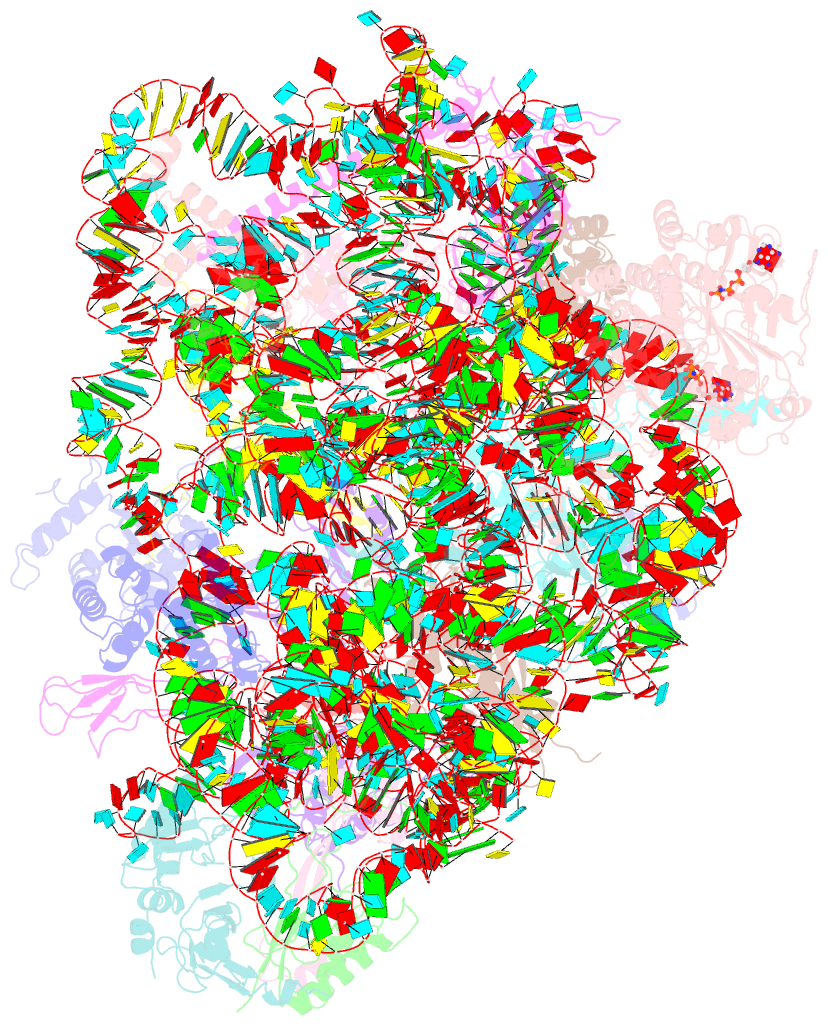
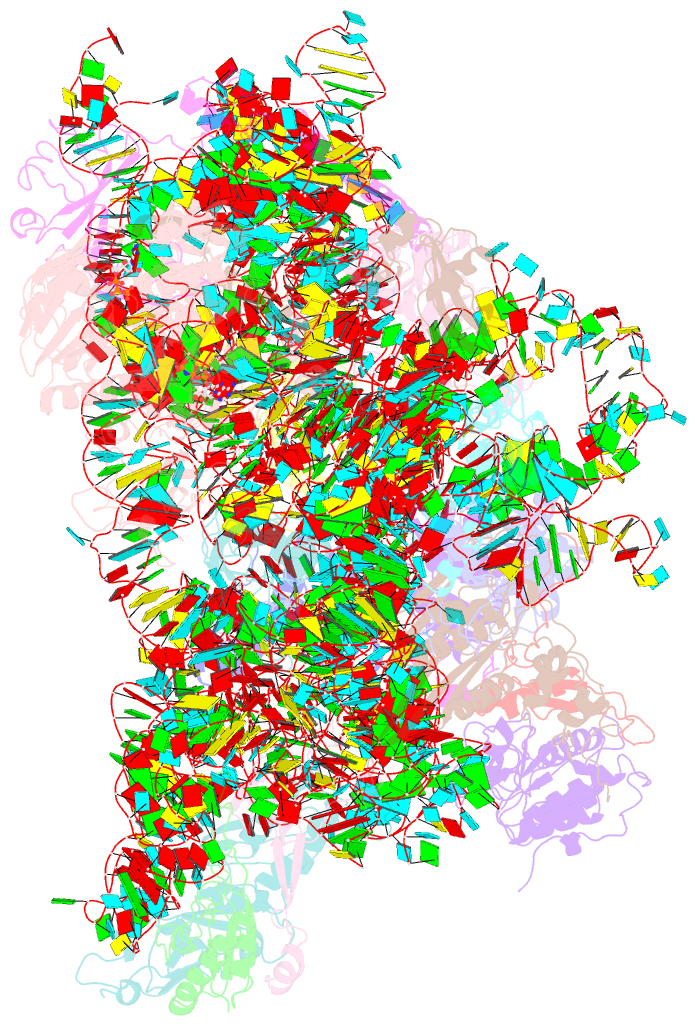
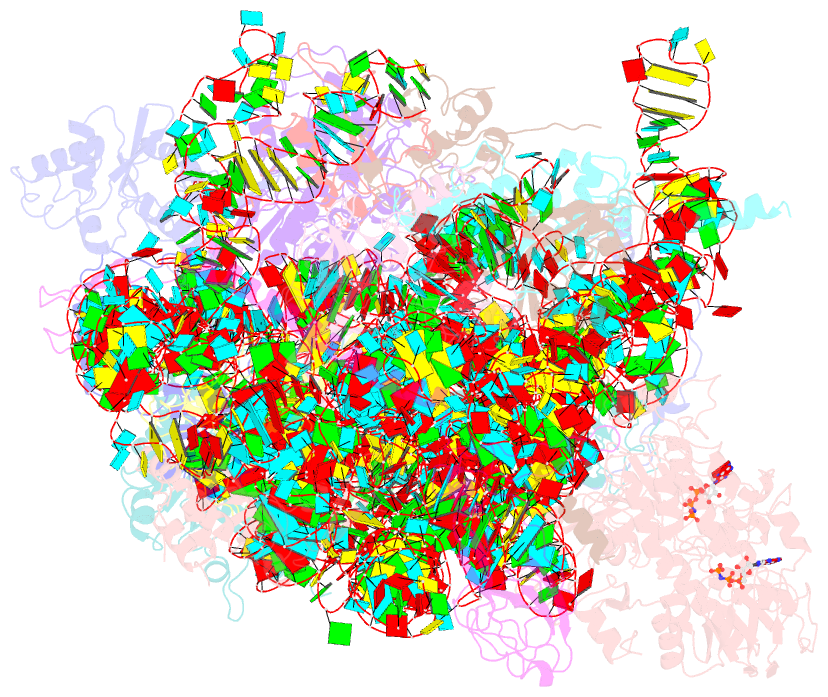
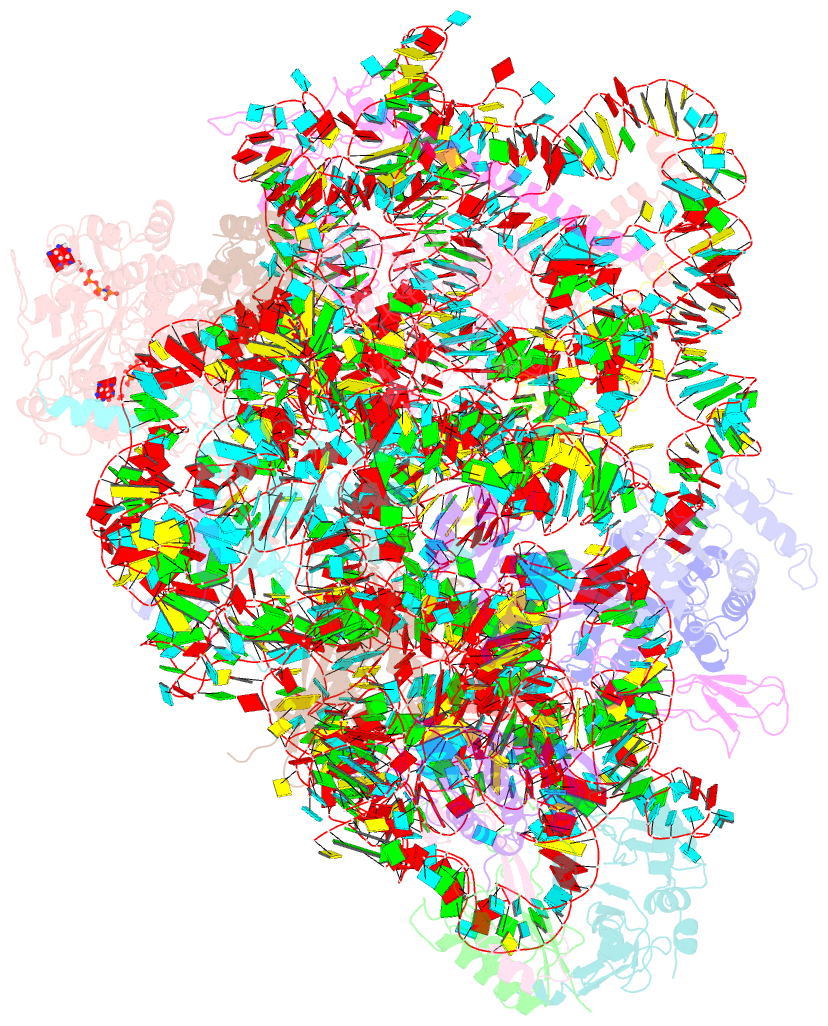
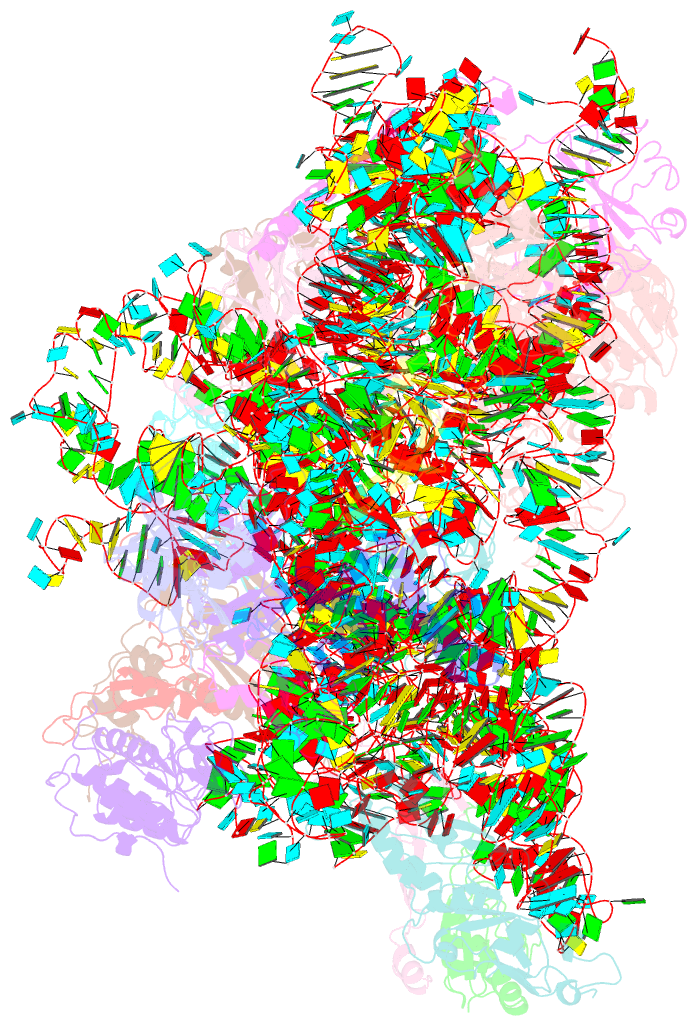
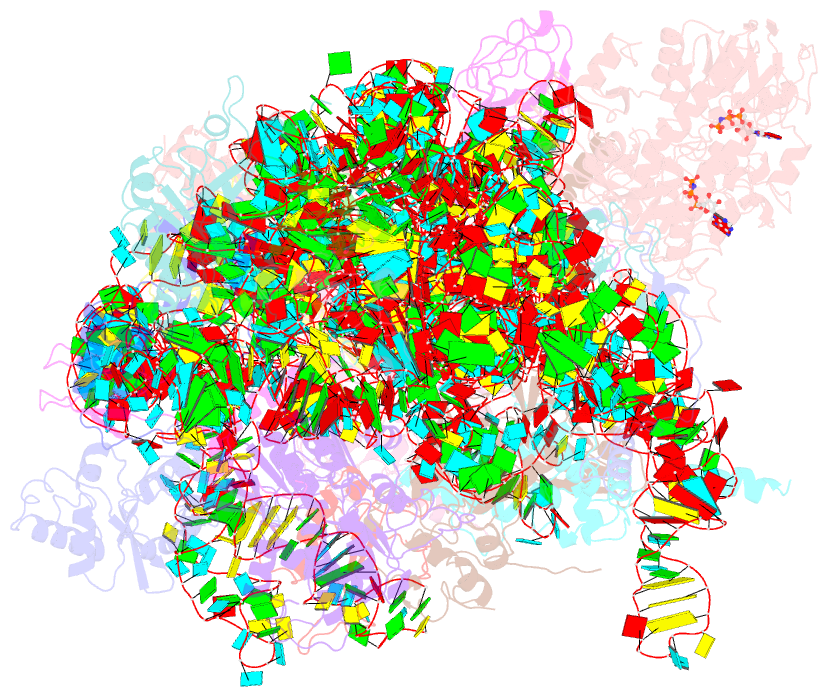
Summary information and primary citation
- PDB-id
- 5ll6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (3.9 Å)
- Summary
- Structure of the 40s abce1 post-splitting complex in ribosome recycling and translation initiation
- Reference
- Heuer A, Gerovac M, Schmidt C, Trowitzsch S, Preis A, Kotter P, Berninghausen O, Becker T, Beckmann R, Tampe R (2017): "Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation." Nat. Struct. Mol. Biol., 24, 453-460. doi: 10.1038/nsmb.3396.
- Abstract
- The essential ATP-binding cassette protein ABCE1 splits 80S ribosomes into 60S and 40S subunits after canonical termination or quality-control-based mRNA surveillance processes. However, the underlying splitting mechanism remains enigmatic. Here, we present a cryo-EM structure of the yeast 40S-ABCE1 post-splitting complex at 3.9-Å resolution. Compared to the pre-splitting state, we observe repositioning of ABCE1's iron-sulfur cluster domain, which rotates 150° into a binding pocket on the 40S subunit. This repositioning explains a newly observed anti-association activity of ABCE1. Notably, the movement implies a collision with A-site factors, thus explaining the splitting mechanism. Disruption of key interactions in the post-splitting complex impairs cellular homeostasis. Additionally, the structure of a native post-splitting complex reveals ABCE1 to be part of the 43S initiation complex, suggesting a coordination of termination, recycling, and initiation.