Summary information and primary citation
- PDB-id
- 5mpf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.918 Å)
- Summary
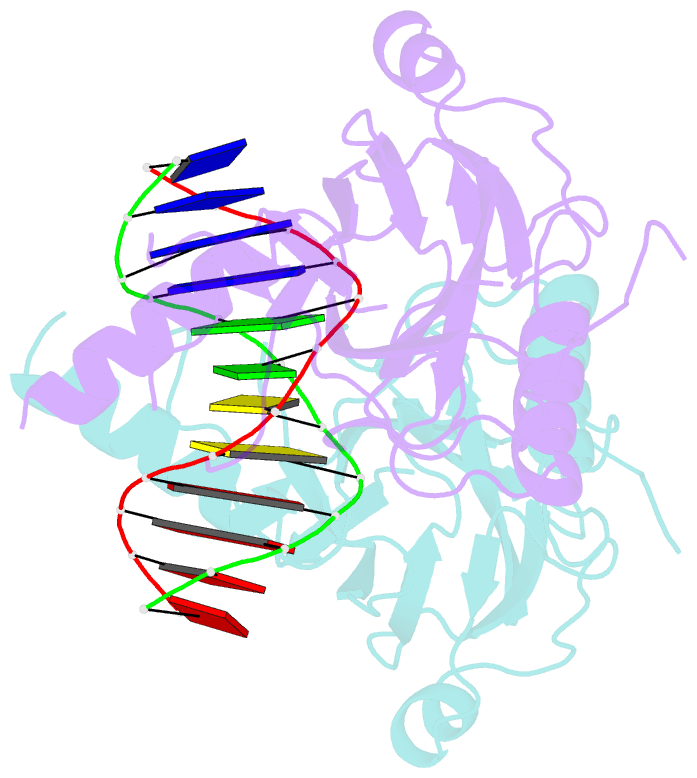
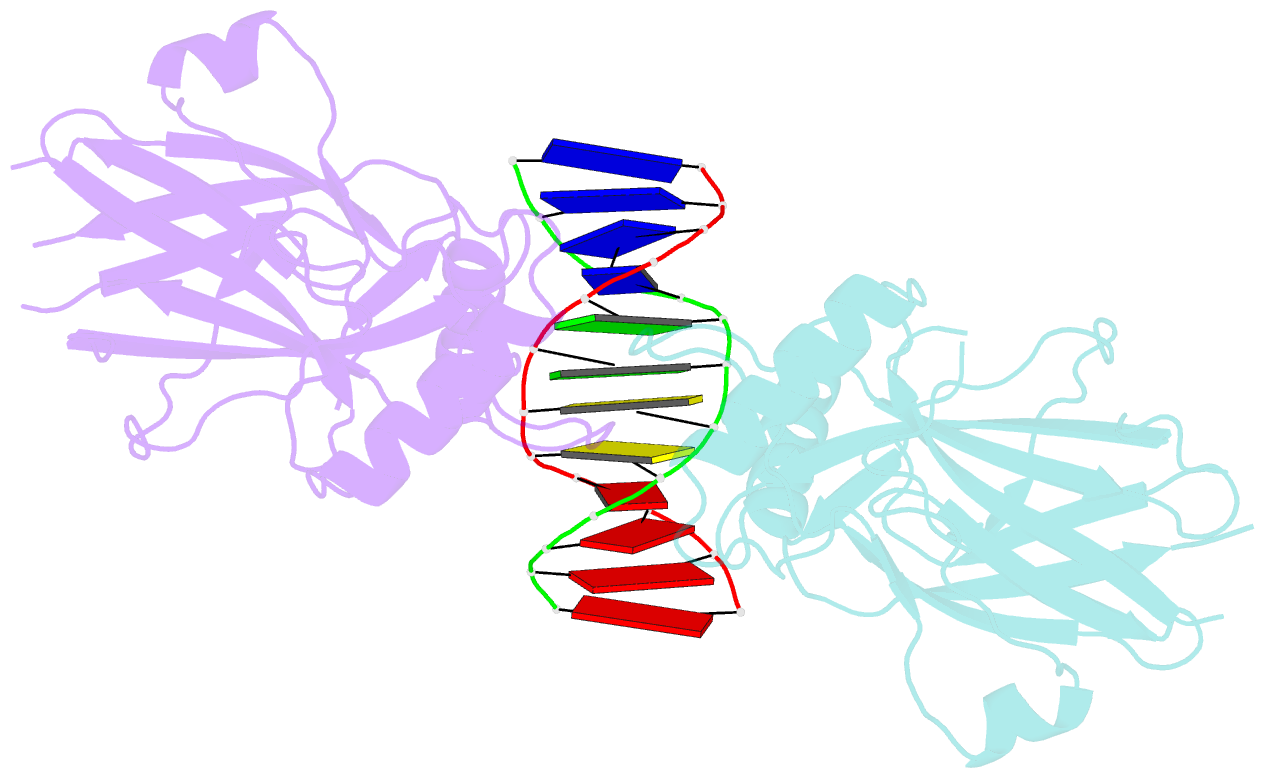
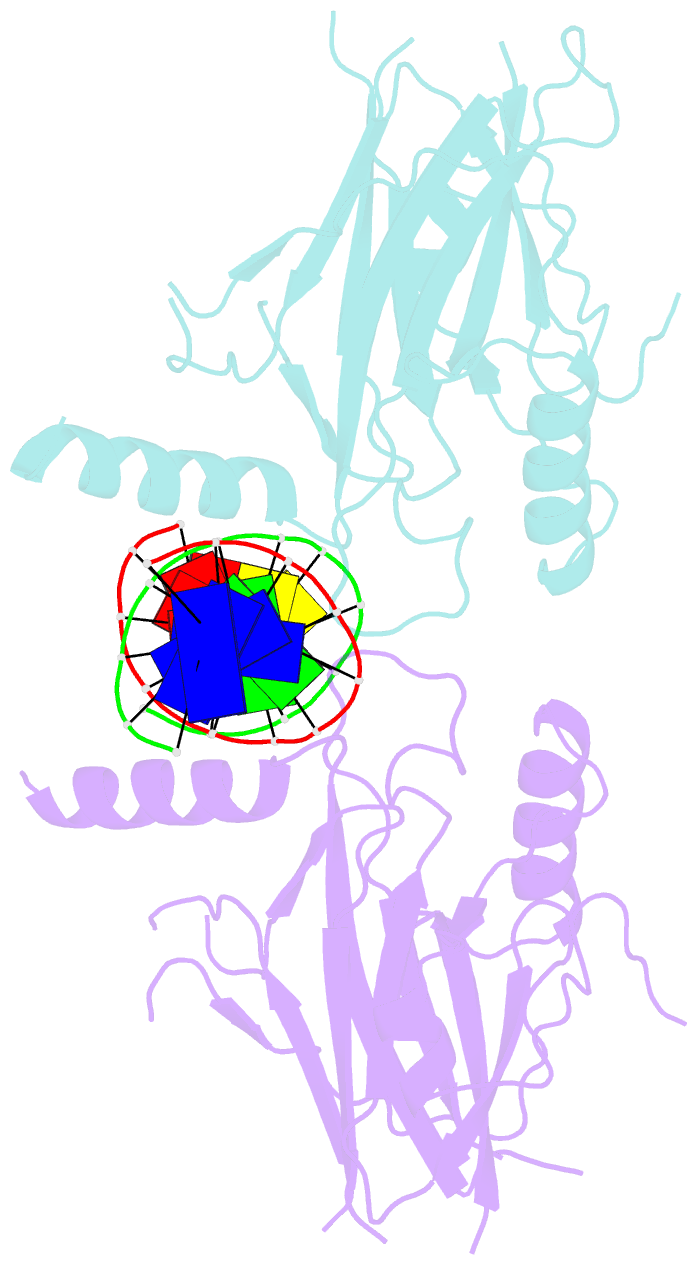
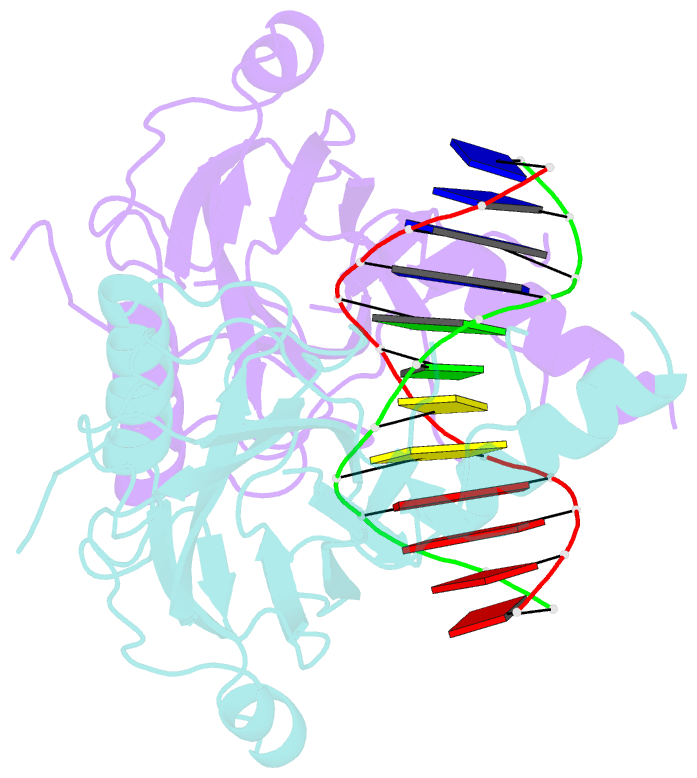
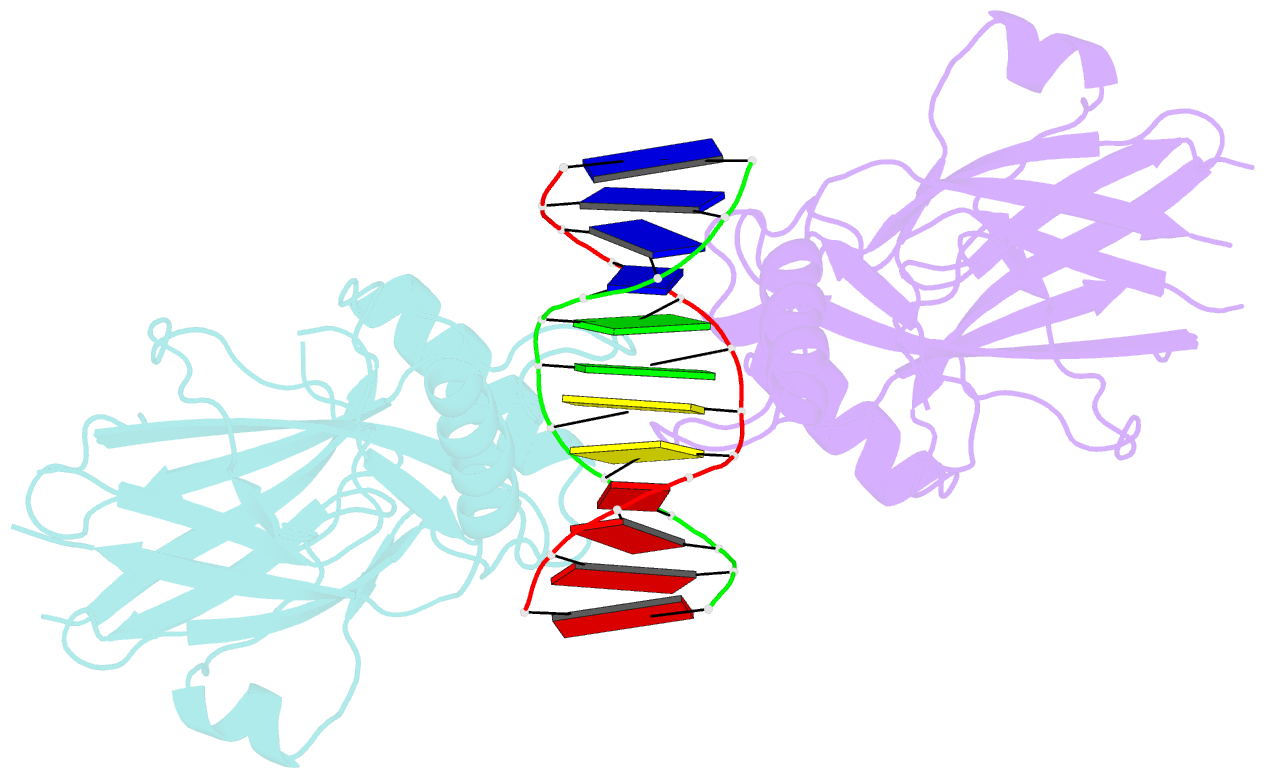
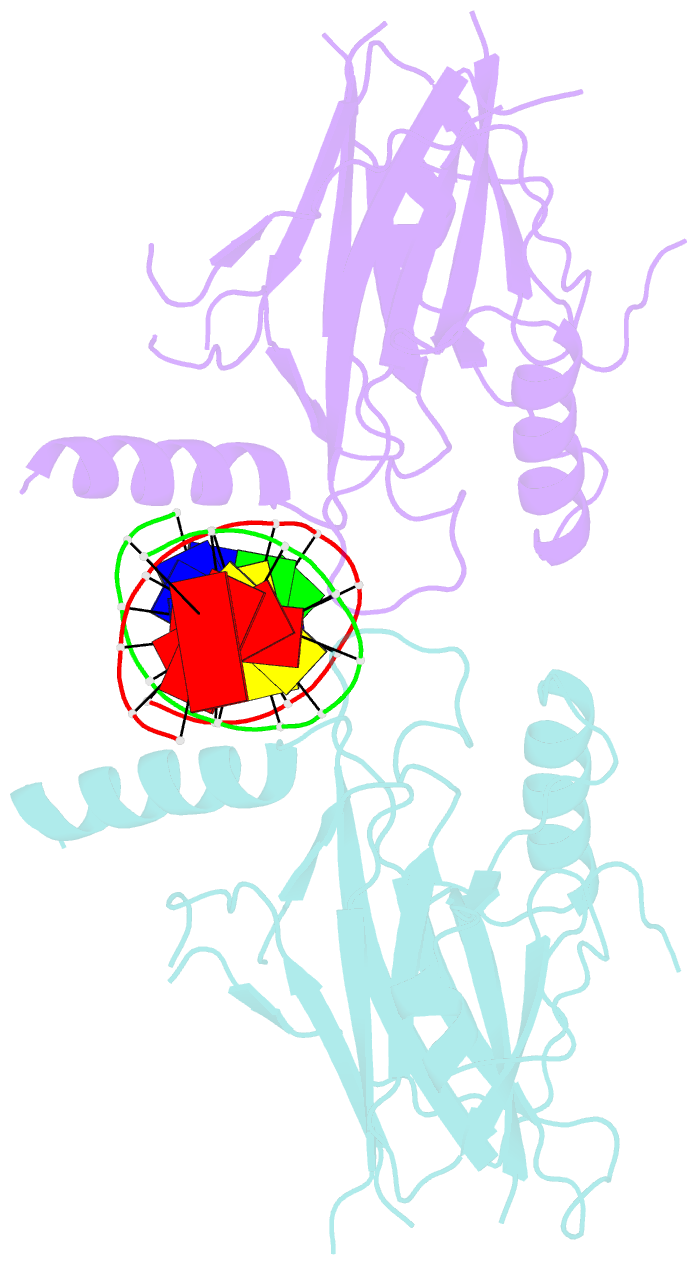
- Structural basis of gene regulation by the grainyhead transcription factor superfamily
- Reference
- Ming Q, Roske Y, Schuetz A, Walentin K, Ibraimi I, Schmidt-Ott KM, Heinemann U (2018): "Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family." Nucleic Acids Res., 46, 2082-2095. doi: 10.1093/nar/gkx1299.
- Abstract
- Grainyhead (Grh)/CP2 transcription factors are highly conserved in multicellular organisms as key regulators of epithelial differentiation, organ development and skin barrier formation. In addition, they have been implicated as being tumor suppressors in a variety of human cancers. Despite their physiological importance, little is known about their structure and DNA binding mode. Here, we report the first structural study of mammalian Grh/CP2 factors. Crystal structures of the DNA-binding domains of grainyhead-like (Grhl) 1 and Grhl2 reveal a closely similar conformation with immunoglobulin-like core. Both share a common fold with the tumor suppressor p53, but differ in important structural features. The Grhl1 DNA-binding domain binds duplex DNA containing the consensus recognition element in a dimeric arrangement, supporting parsimonious target-sequence selection through two conserved arginine residues. We elucidate the molecular basis of a cancer-related mutation in Grhl1 involving one of these arginines, which completely abrogates DNA binding in biochemical assays and transcriptional activation of a reporter gene in a human cell line. Thus, our studies establish the structural basis of DNA target-site recognition by Grh transcription factors and reveal how tumor-associated mutations inactivate Grhl proteins. They may serve as points of departure for the structure-based development of Grh/CP2 inhibitors for therapeutic applications.