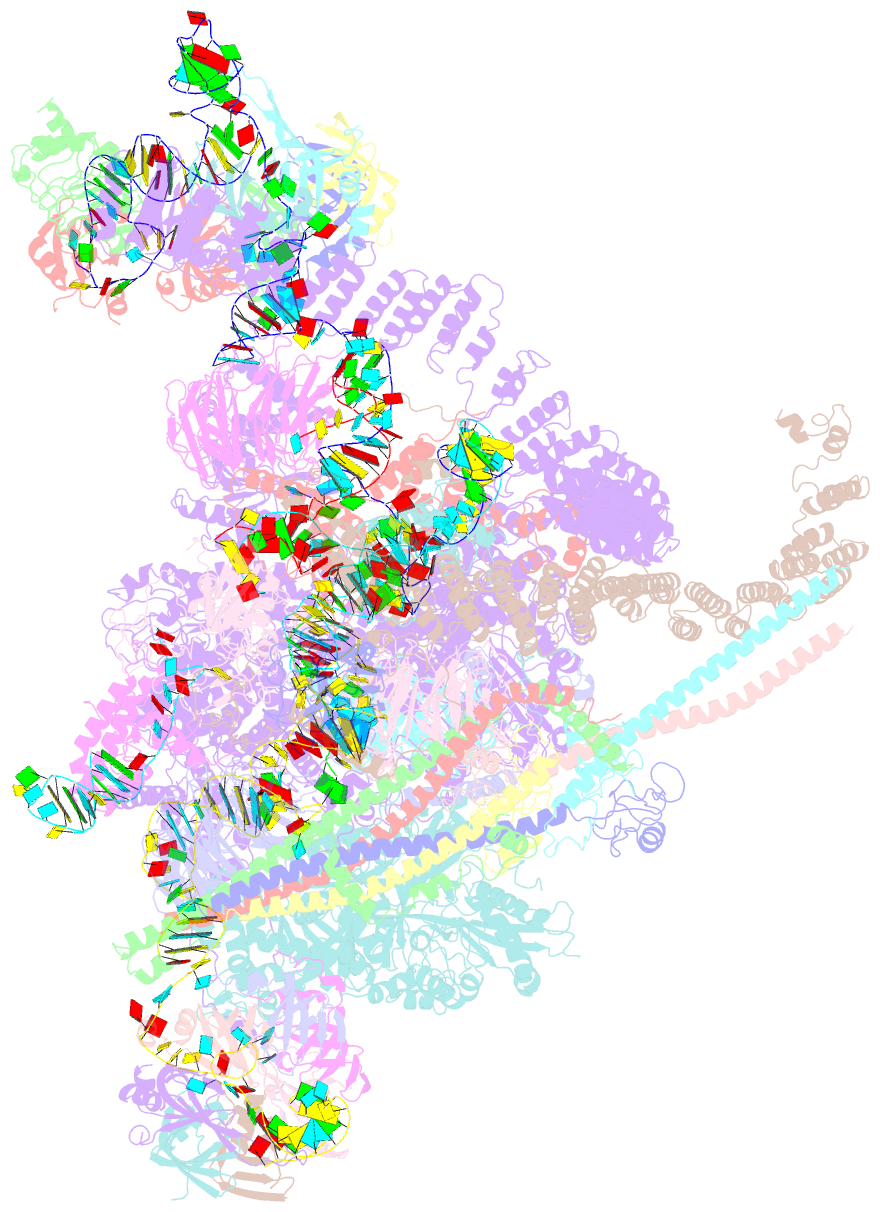
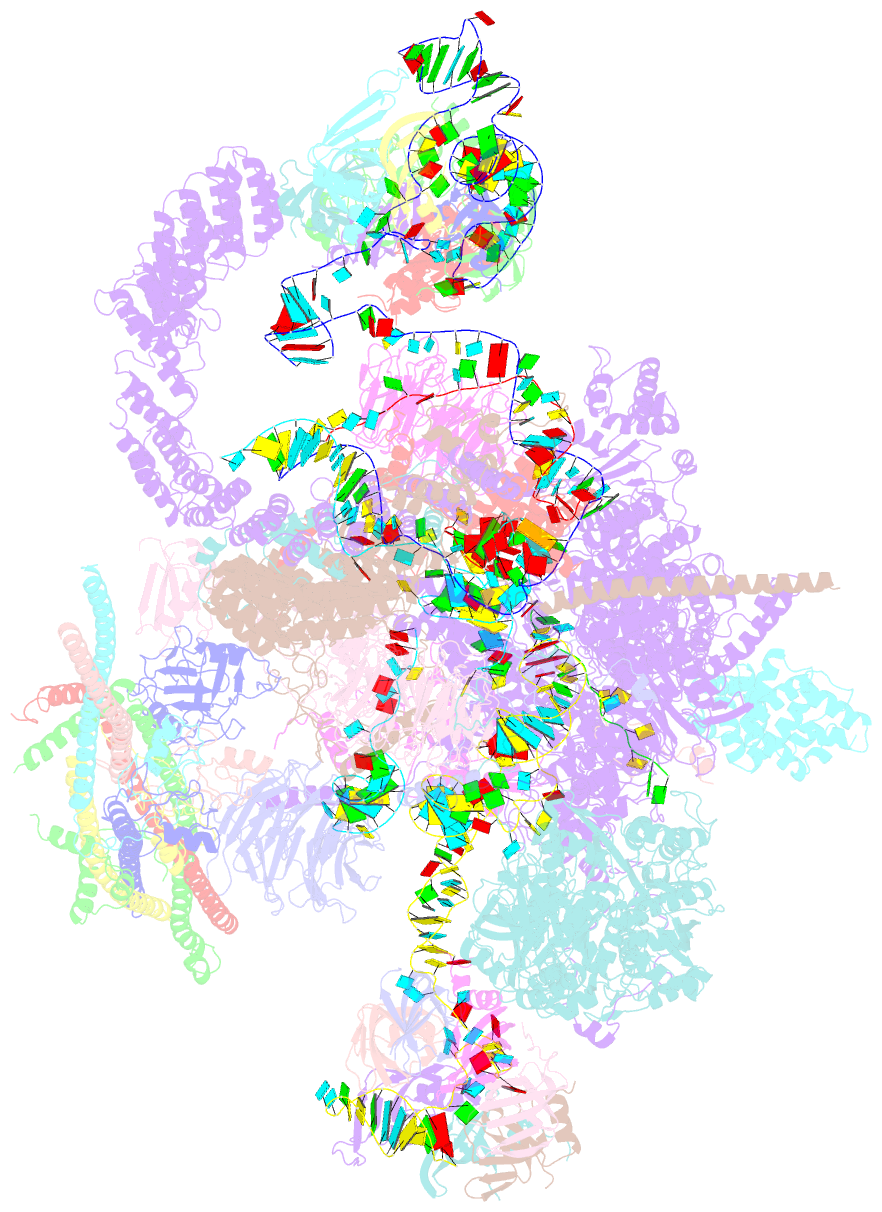
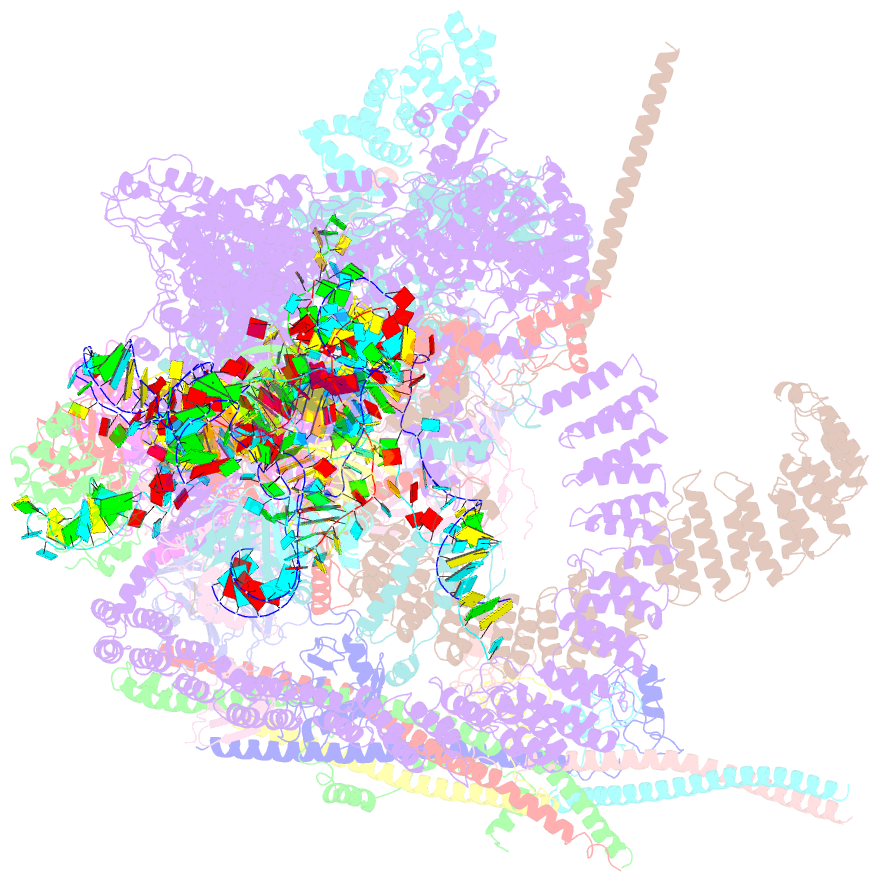
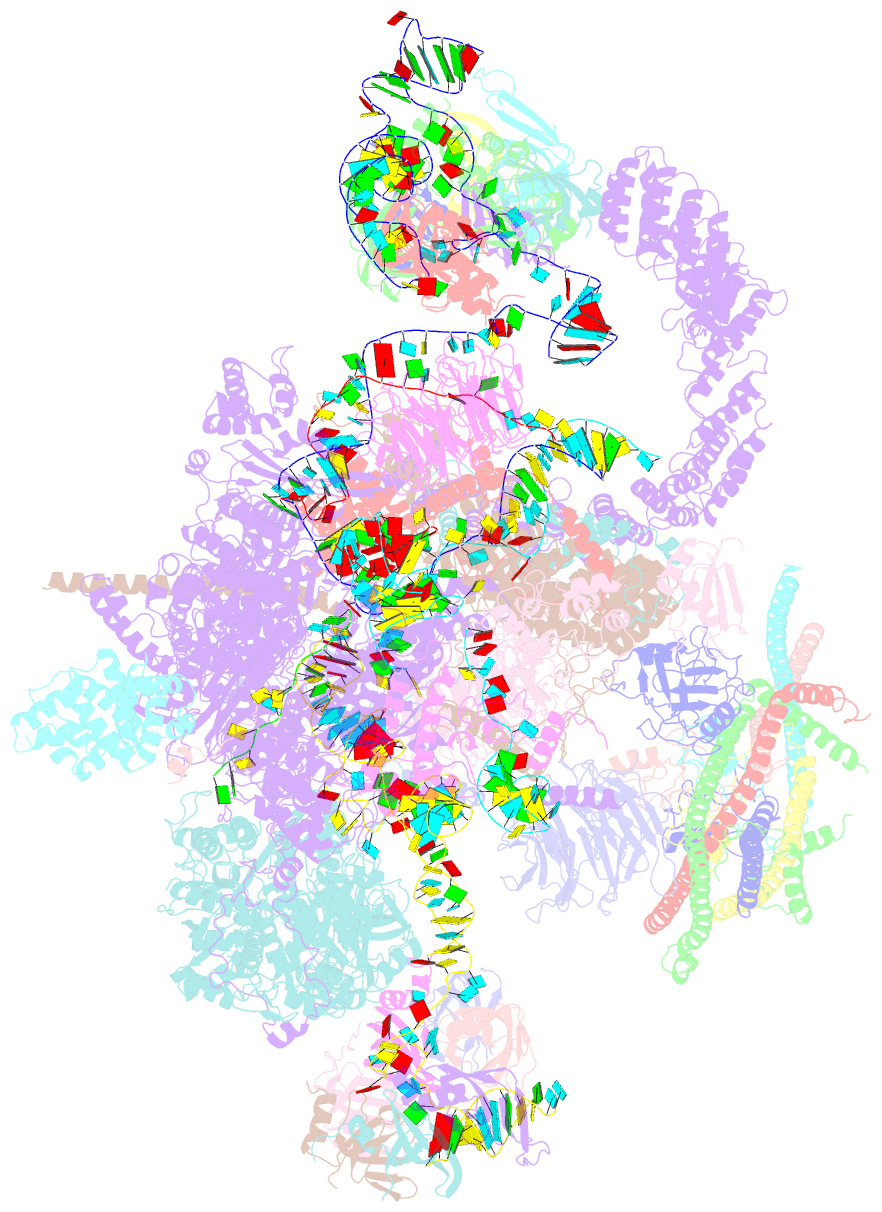
Summary information and primary citation
- PDB-id
- 5mqf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- cryo-EM (5.9 Å)
- Summary
- cryo-EM structure of a human spliceosome activated for step 2 of splicing (c* complex)
- Reference
- Bertram K, Agafonov DE, Liu WT, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lu Hrmann R (2017): "Cryo-EM structure of a human spliceosome activated for step 2 of splicing." Nature, 542, 318-323. doi: 10.1038/nature21079.
- Abstract
- Spliceosome rearrangements facilitated by RNA helicase PRP16 before catalytic step two of splicing are poorly understood. Here we report a 3D cryo-electron microscopy structure of the human spliceosomal C complex stalled directly after PRP16 action (C*). The architecture of the catalytic U2-U6 ribonucleoprotein (RNP) core of the human C* spliceosome is very similar to that of the yeast pre-Prp16 C complex. However, in C* the branched intron region is separated from the catalytic centre by approximately 20 Å, and its position close to the U6 small nuclear RNA ACAGA box is stabilized by interactions with the PRP8 RNase H-like and PRP17 WD40 domains. RNA helicase PRP22 is located about 100 Å from the catalytic centre, suggesting that it destabilizes the spliced mRNA after step two from a distance. Comparison of the structure of the yeast C and human C* complexes reveals numerous RNP rearrangements that are likely to be facilitated by PRP16, including a large-scale movement of the U2 small nuclear RNP.