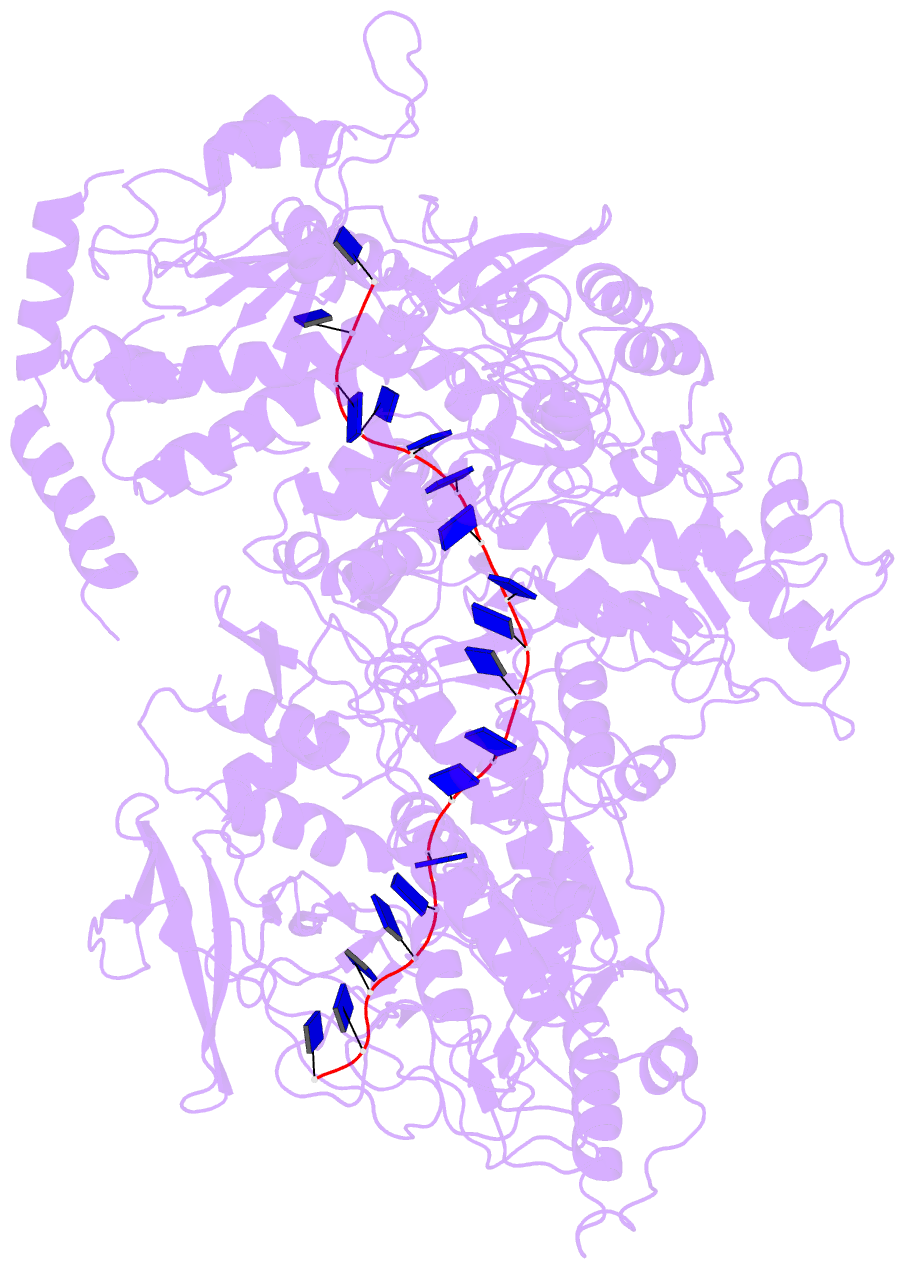
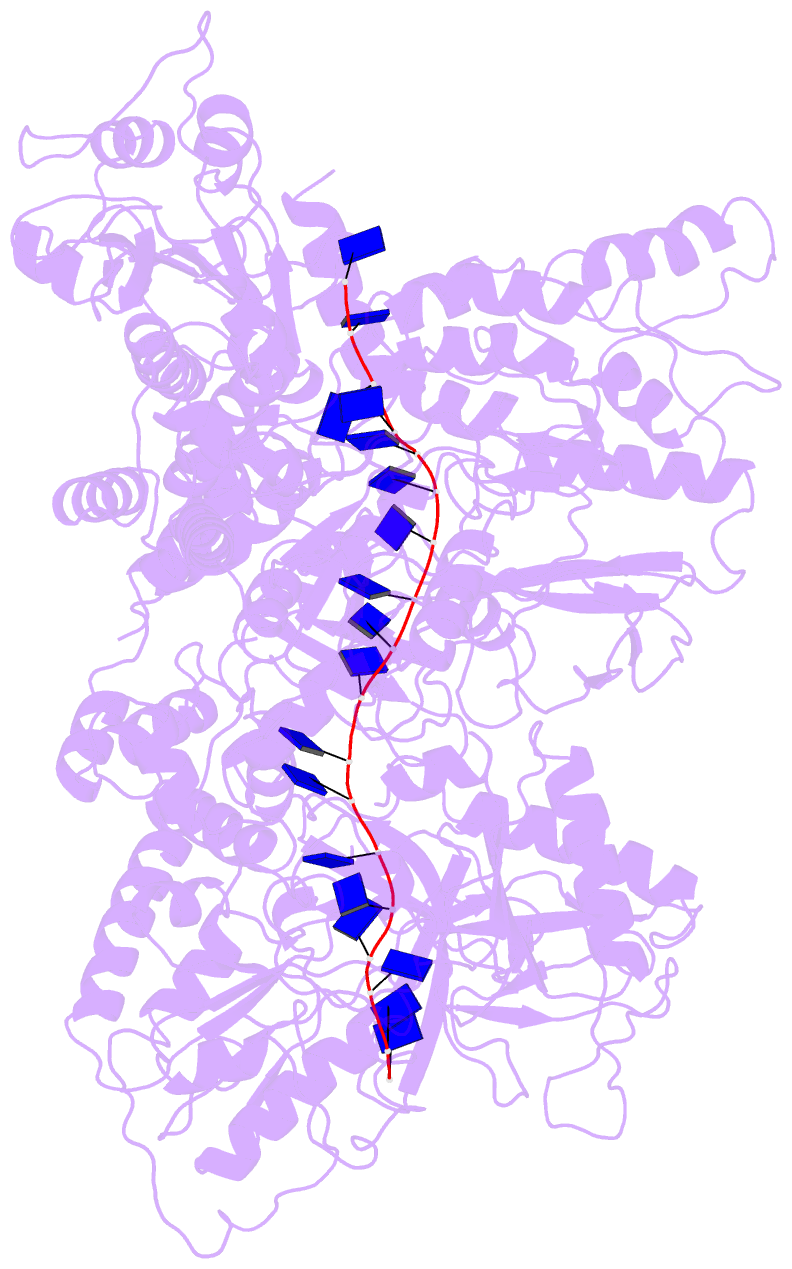
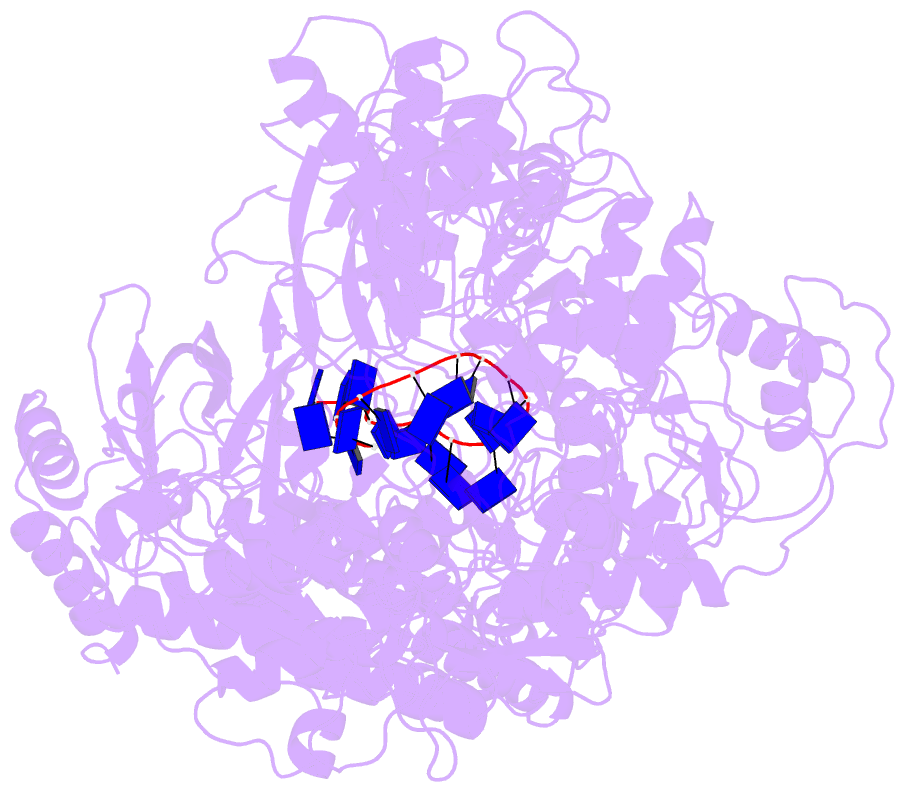
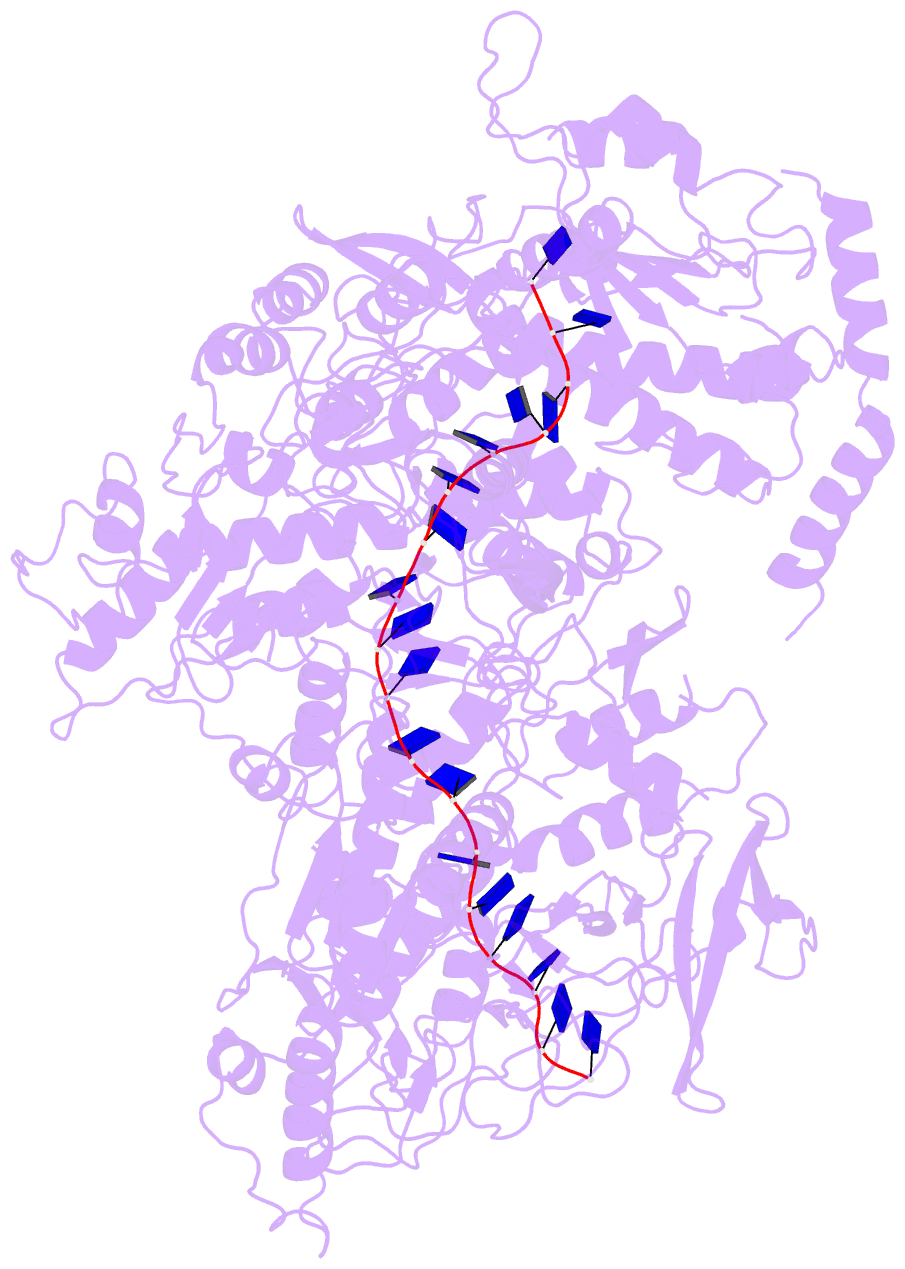
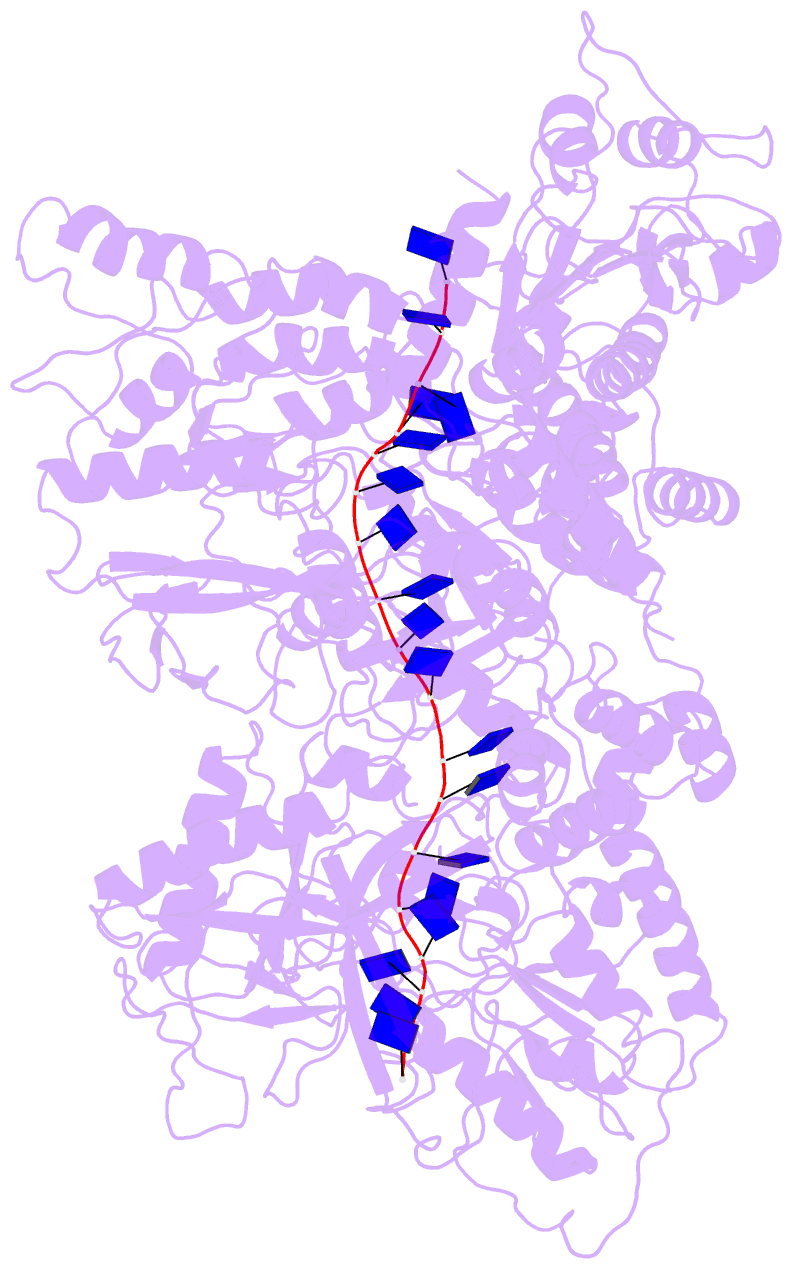
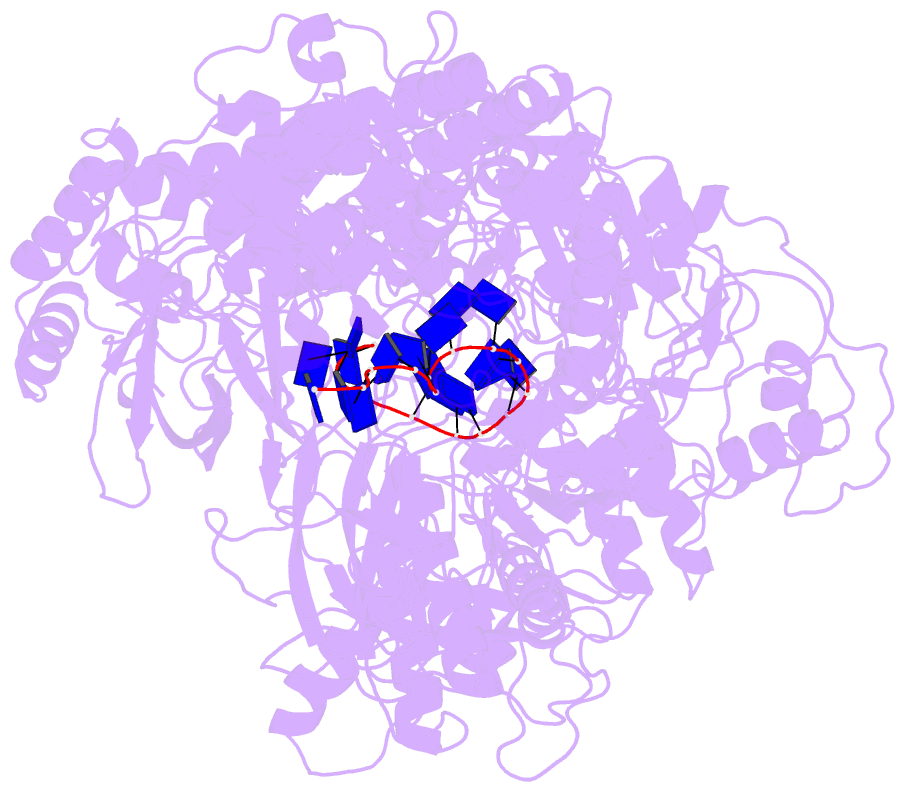
Summary information and primary citation
- PDB-id
- 5n8o; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- cryo-EM (3.9 Å)
- Summary
- Cryo em structure of the conjugative relaxase trai of the f-r1 plasmid system
- Reference
- Ilangovan A, Kay CWM, Roier S, El Mkami H, Salvadori E, Zechner EL, Zanetti G, Waksman G (2017): "Cryo-EM Structure of a Relaxase Reveals the Molecular Basis of DNA Unwinding during Bacterial Conjugation." Cell, 169, 708-721.e12. doi: 10.1016/j.cell.2017.04.010.
- Abstract
- Relaxases play essential roles in conjugation, the main process by which bacteria exchange genetic material, notably antibiotic resistance genes. They are bifunctional enzymes containing a trans-esterase activity, which is responsible for nicking the DNA strand to be transferred and for covalent attachment to the resulting 5'-phosphate end, and a helicase activity, which is responsible for unwinding the DNA while it is being transported to a recipient cell. Here we show that these two activities are carried out by two conformers that can both load simultaneously on the origin of transfer DNA. We solve the structure of one of these conformers by cryo electron microscopy to near-atomic resolution, elucidating the molecular basis of helicase function by relaxases and revealing insights into the mechanistic events taking place in the cell prior to substrate transport during conjugation.