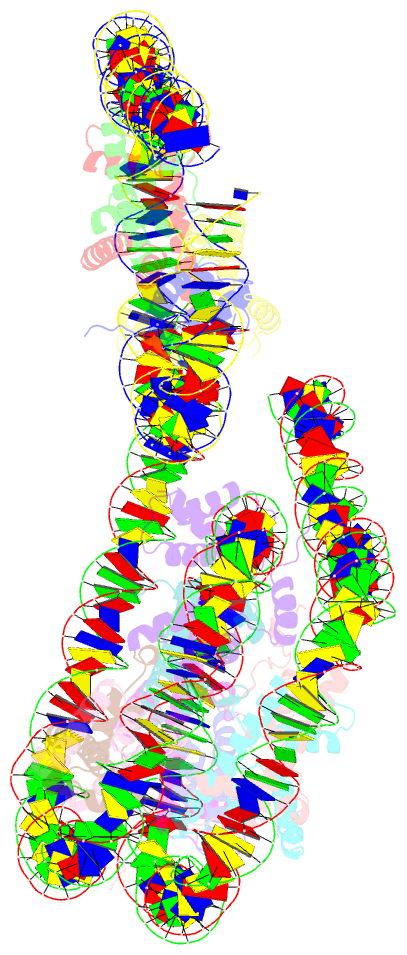
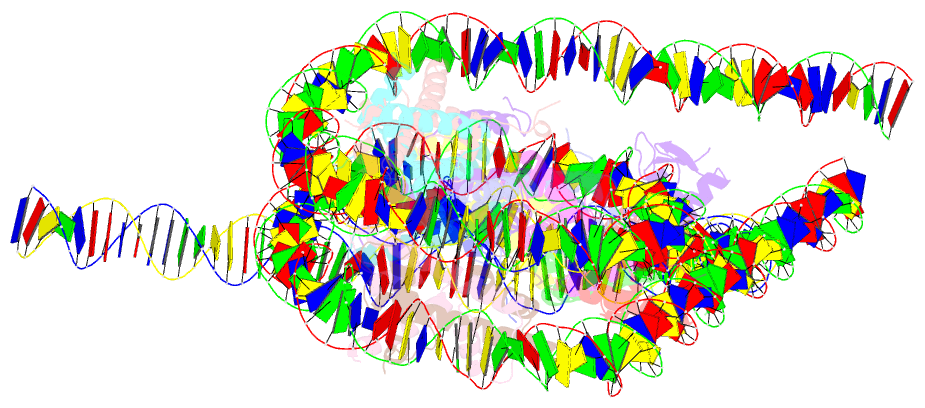
Summary information and primary citation
- PDB-id
- 5nl0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- chromatin binding protein - DNA
- Method
- X-ray (5.4 Å)
- Summary
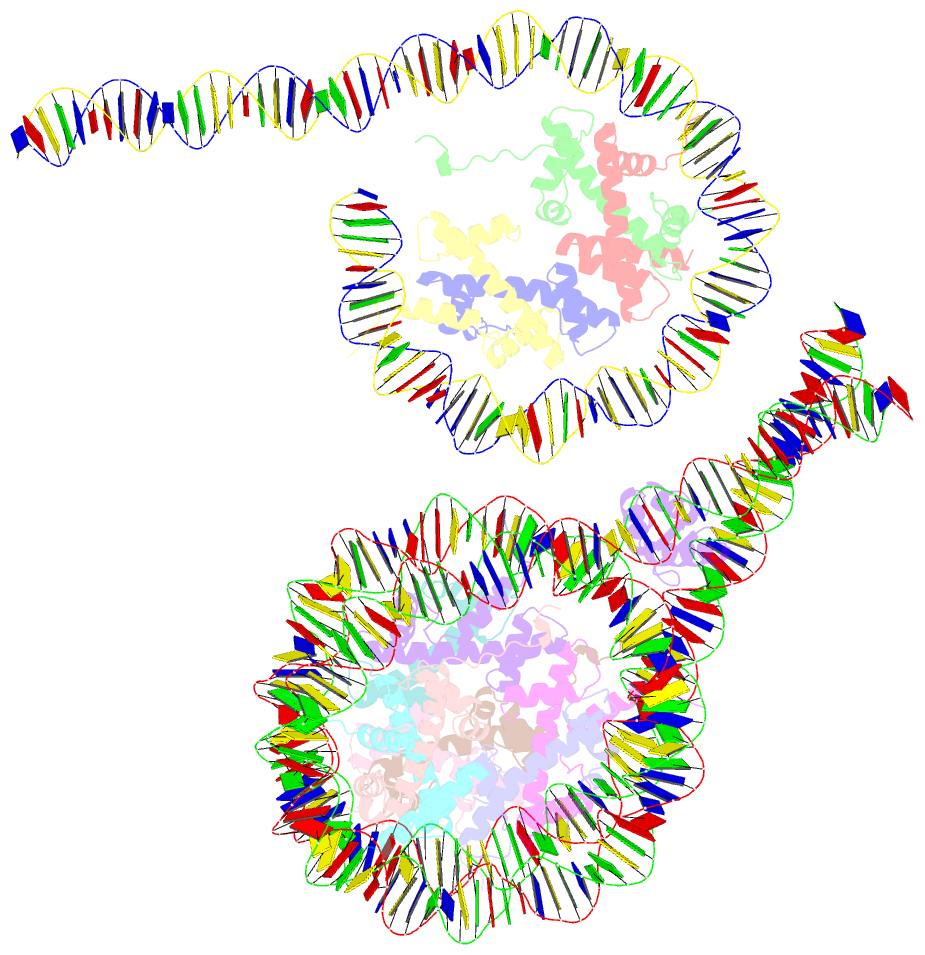
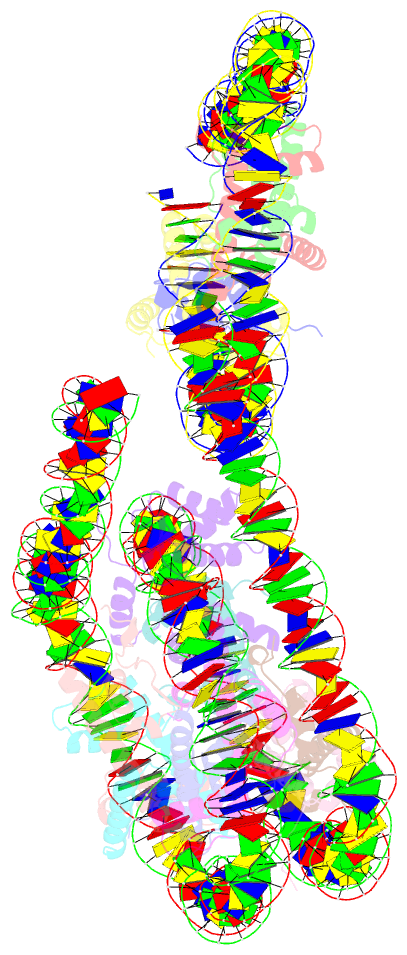
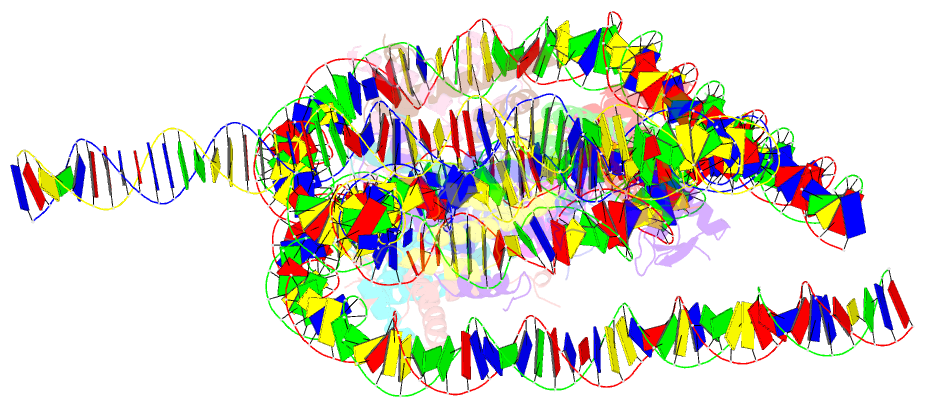
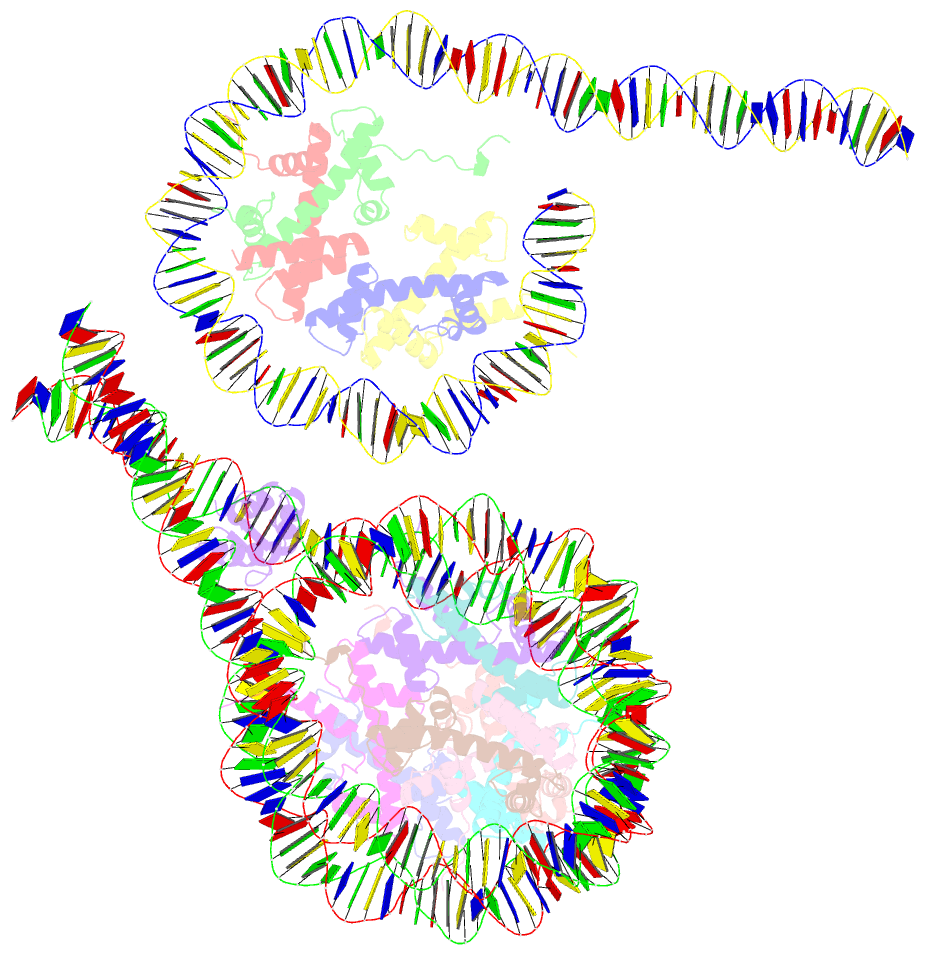
- Crystal structure of a 197-bp palindromic 601l nucleosome in complex with linker histone h1
- Reference
- Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Papai G, Reymer A, Syed SH, Lone IN, Tonchev O, Crucifix C, Menoni H, Papin C, Skoufias DA, Kurumizaka H, Lavery R, Hamiche A, Hayes JJ, Schultz P, Angelov D, Petosa C, Dimitrov S (2017): "Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1." Mol. Cell, 66, 384-397.e8. doi: 10.1016/j.molcel.2017.04.012.
- Abstract
- Linker histones associate with nucleosomes to promote the formation of higher-order chromatin structure, but the underlying molecular details are unclear. We investigated the structure of a 197 bp nucleosome bearing symmetric 25 bp linker DNA arms in complex with vertebrate linker histone H1. We determined electron cryo-microscopy (cryo-EM) and crystal structures of unbound and H1-bound nucleosomes and validated these structures by site-directed protein cross-linking and hydroxyl radical footprinting experiments. Histone H1 shifts the conformational landscape of the nucleosome by drawing the two linkers together and reducing their flexibility. The H1 C-terminal domain (CTD) localizes primarily to a single linker, while the H1 globular domain contacts the nucleosome dyad and both linkers, associating more closely with the CTD-distal linker. These findings reveal that H1 imparts a strong degree of asymmetry to the nucleosome, which is likely to influence the assembly and architecture of higher-order structures.