Summary information and primary citation
- PDB-id
- 5nwa; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (3.2 Å)
- Summary
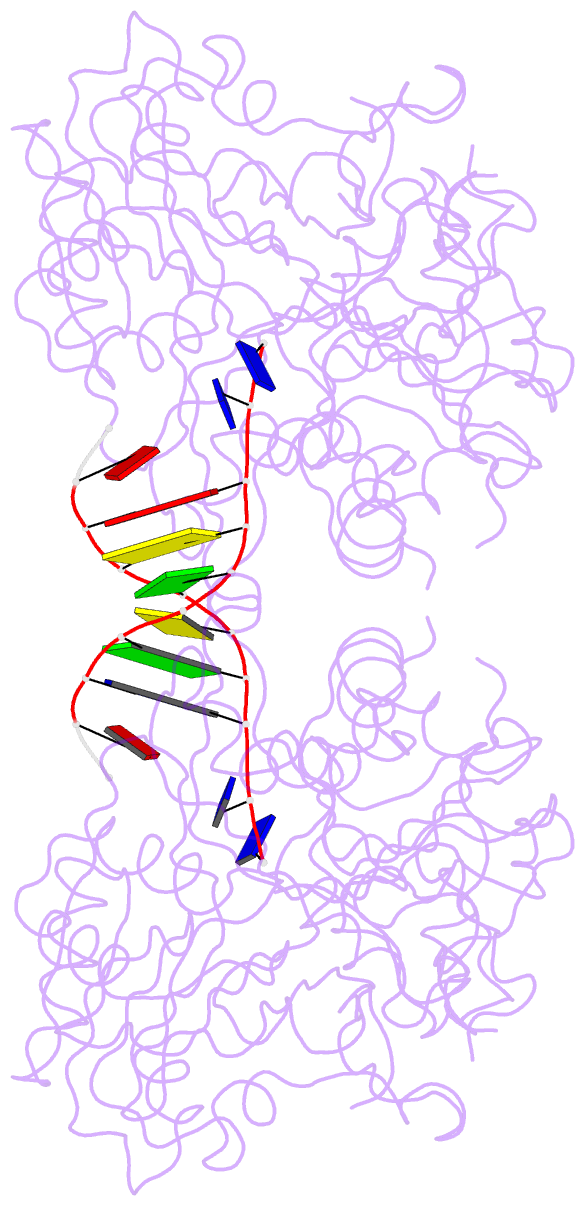
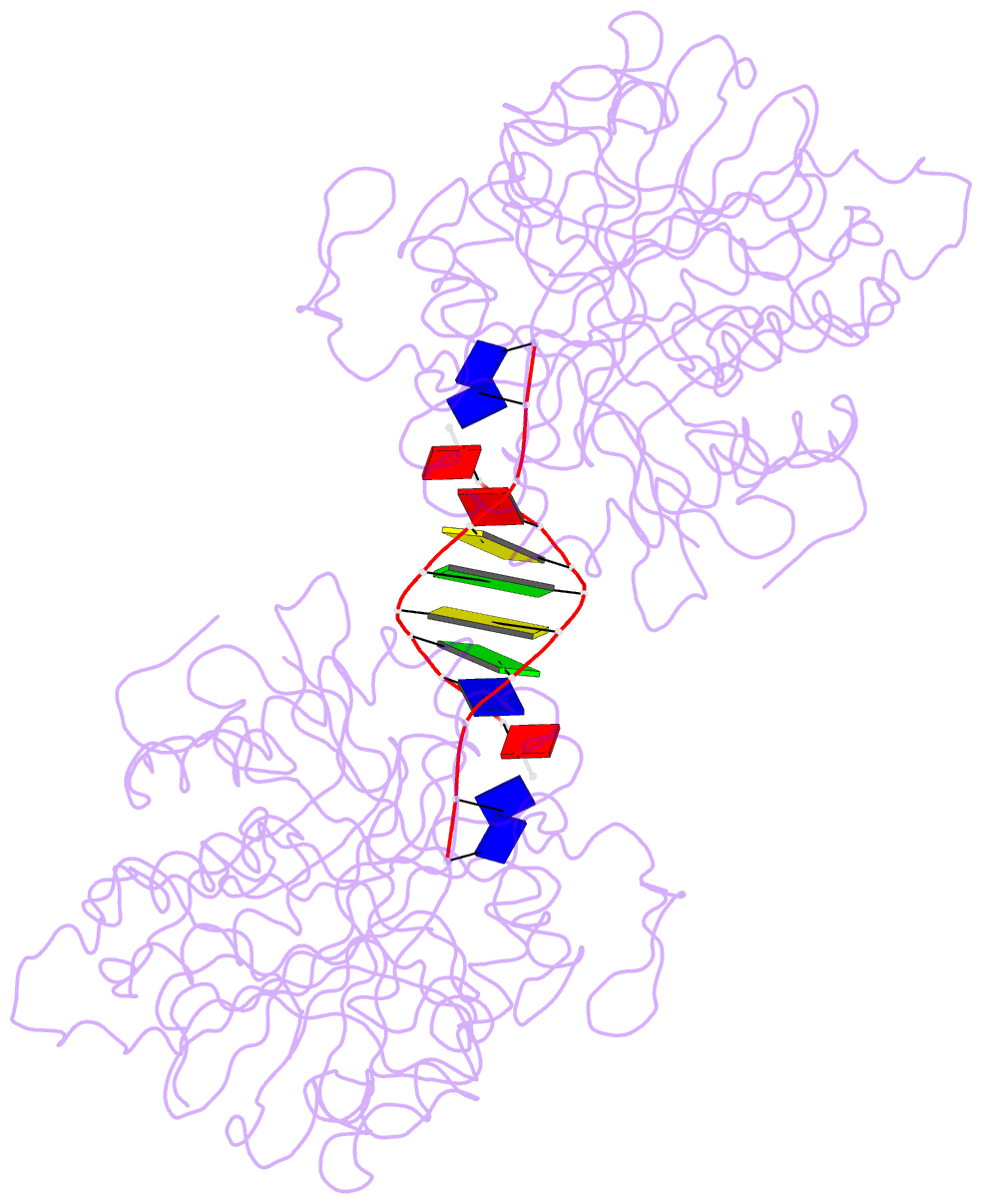
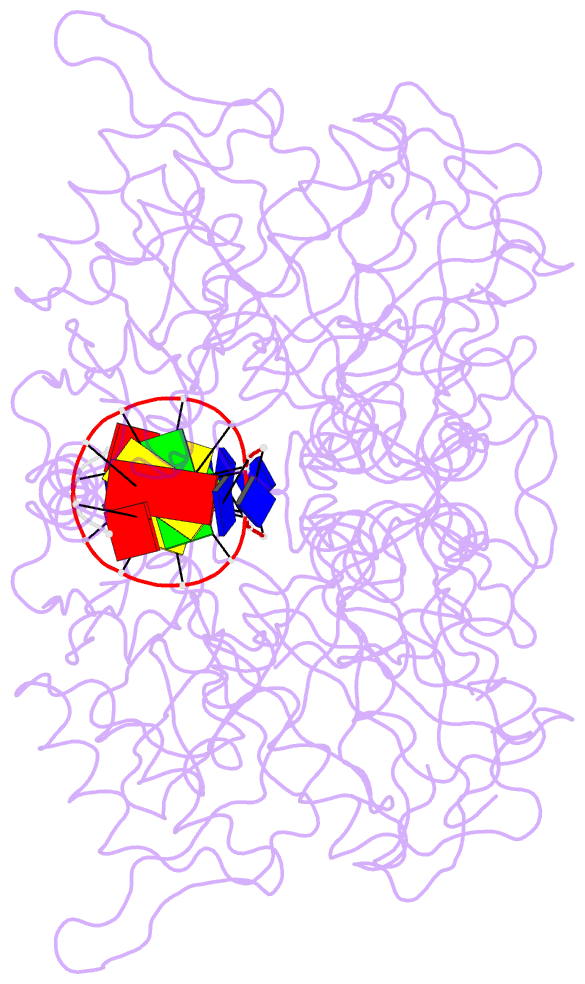
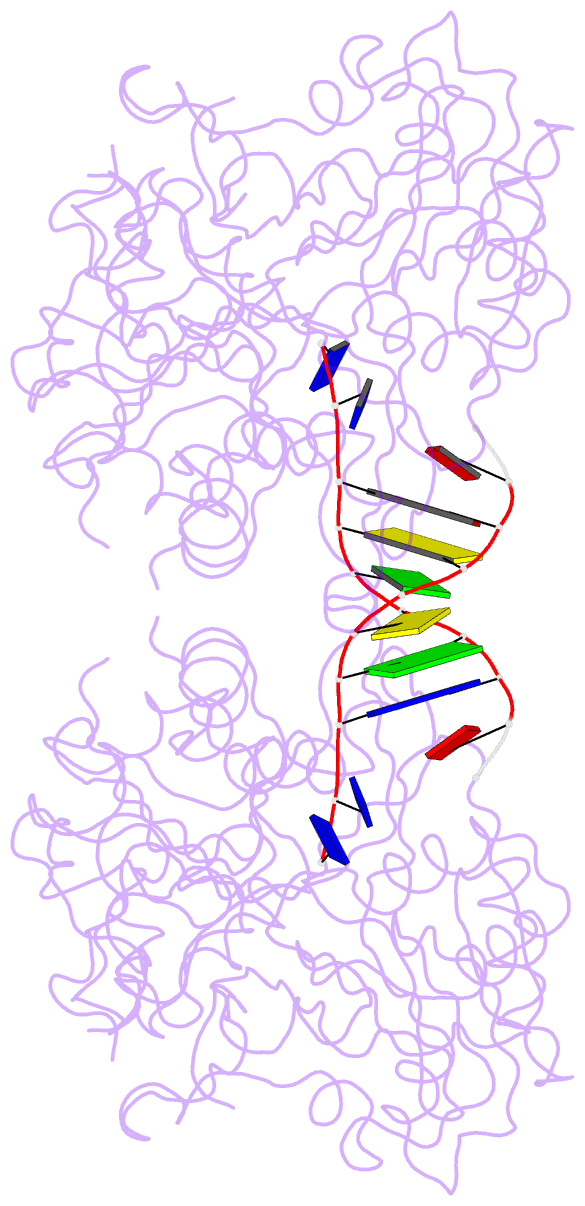
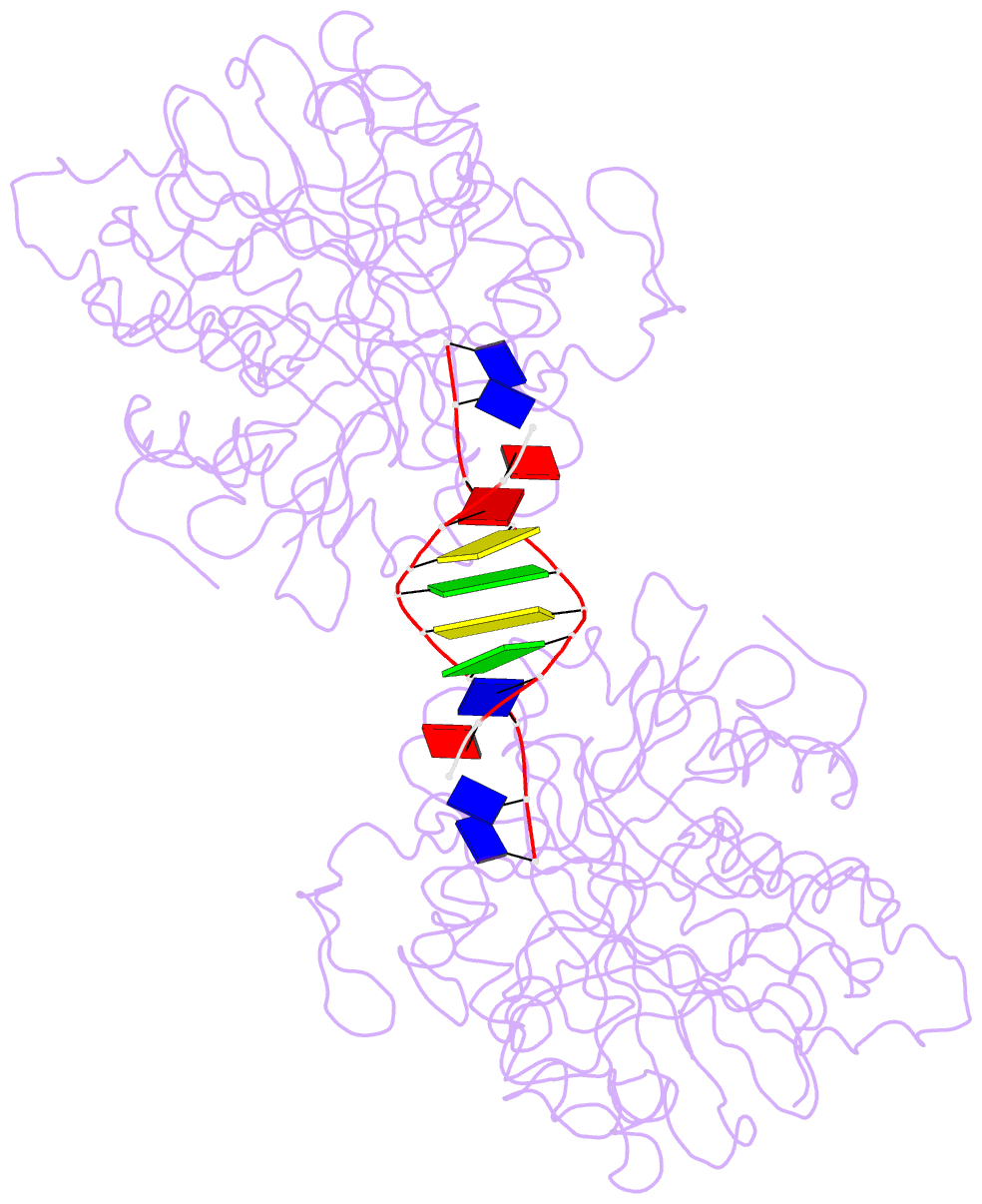
- Crystal structure of the complex of tdp1 with duplex DNA
- Reference
- Flett FJ, Ruksenaite E, Armstrong LA, Bharati S, Carloni R, Morris ER, Mackay CL, Interthal H, Richardson JM (2018): "Structural basis for DNA 3'-end processing by human tyrosyl-DNA phosphodiesterase 1." Nat Commun, 9, 24. doi: 10.1038/s41467-017-02530-z.
- Abstract
- Tyrosyl-DNA phosphodiesterase (Tdp1) is a DNA 3'-end processing enzyme that repairs topoisomerase 1B-induced DNA damage. We use a new tool combining site-specific DNA-protein cross-linking with mass spectrometry to identify Tdp1 interactions with DNA. A conserved phenylalanine (F259) of Tdp1, required for efficient DNA processing in biochemical assays, cross-links to defined positions in DNA substrates. Crystal structures of Tdp1-DNA complexes capture the DNA repair machinery after 3'-end cleavage; these reveal how Tdp1 coordinates the 3'-phosphorylated product of nucleosidase activity and accommodates duplex DNA. A hydrophobic wedge splits the DNA ends, directing the scissile strand through a channel towards the active site. The F259 side-chain stacks against the -3 base pair, delimiting the junction of duplexed and melted DNA, and fixes the scissile strand in the channel. Our results explain why Tdp1 cleavage is non-processive and provide a molecular basis for DNA 3'-end processing by Tdp1.