Summary information and primary citation
- PDB-id
- 5o3j; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- X-ray (2.97 Å)
- Summary
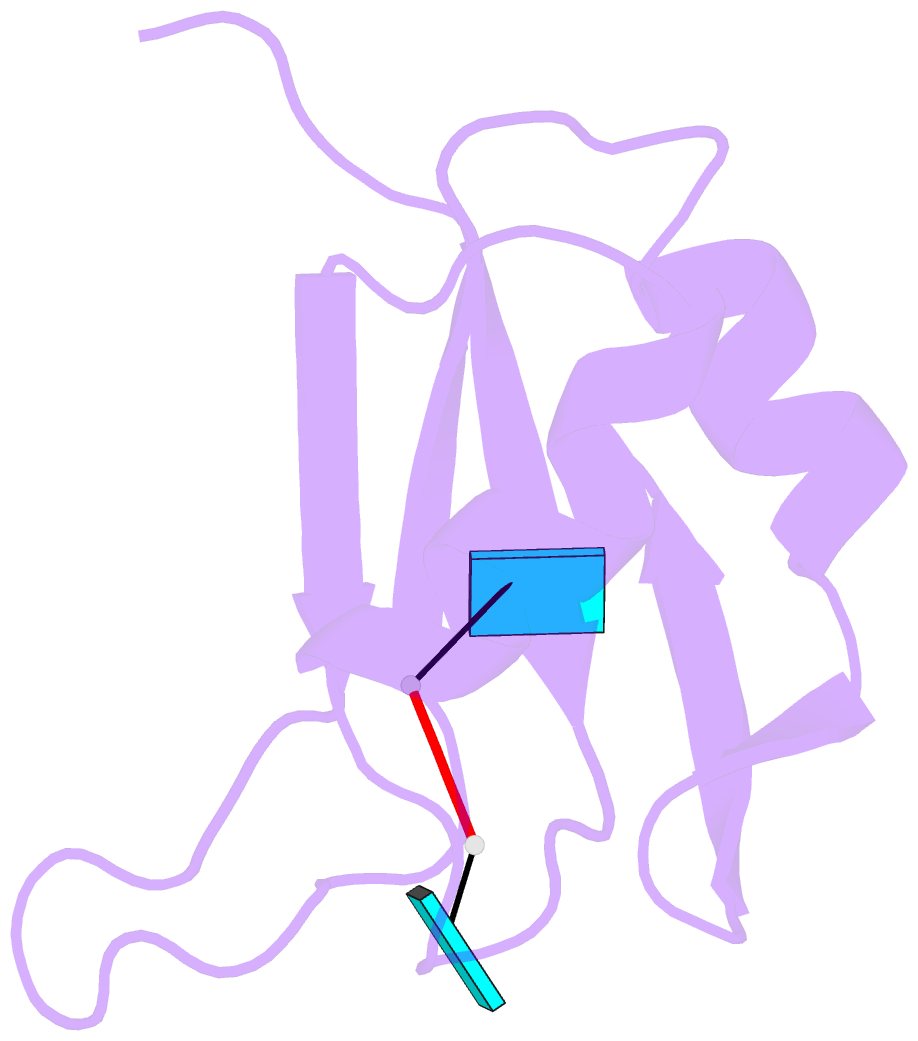
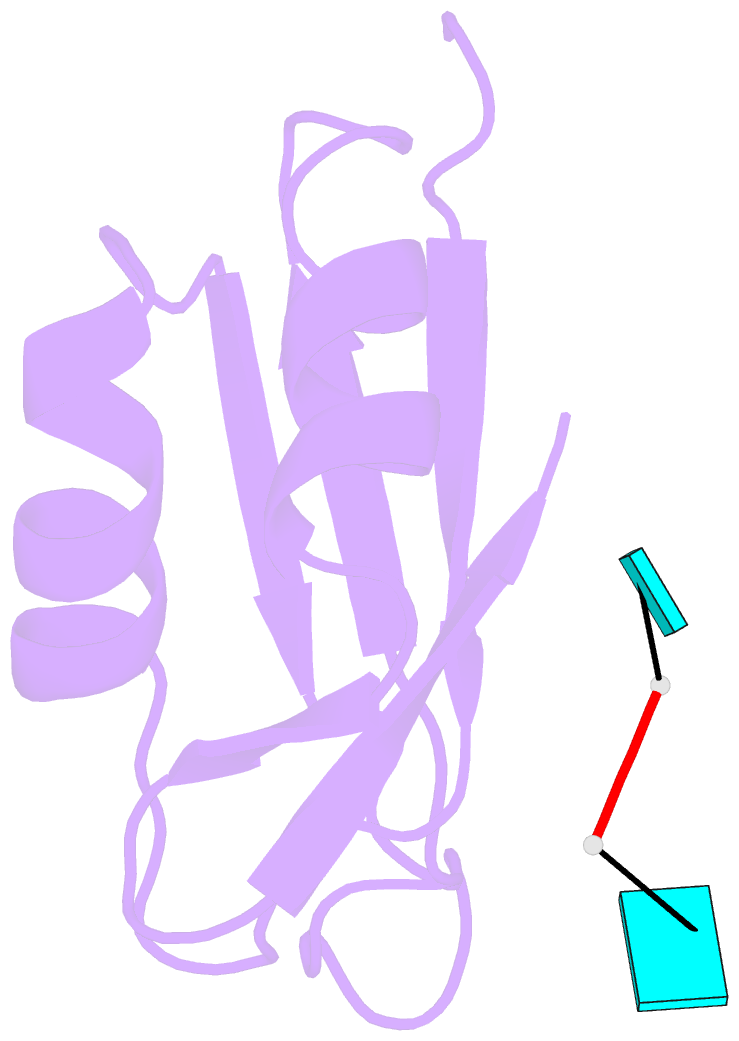
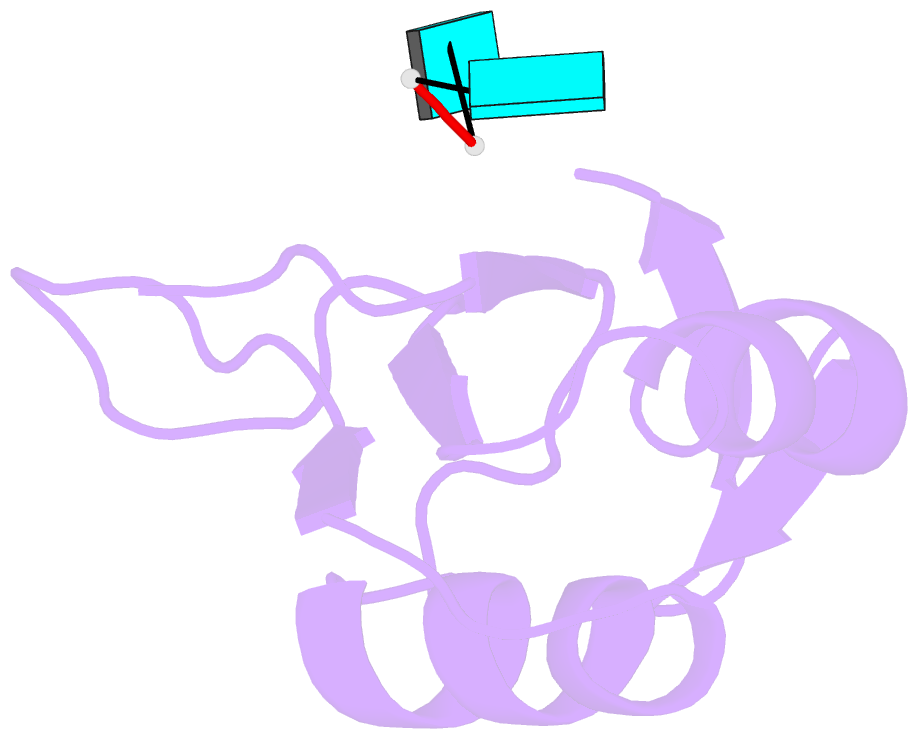
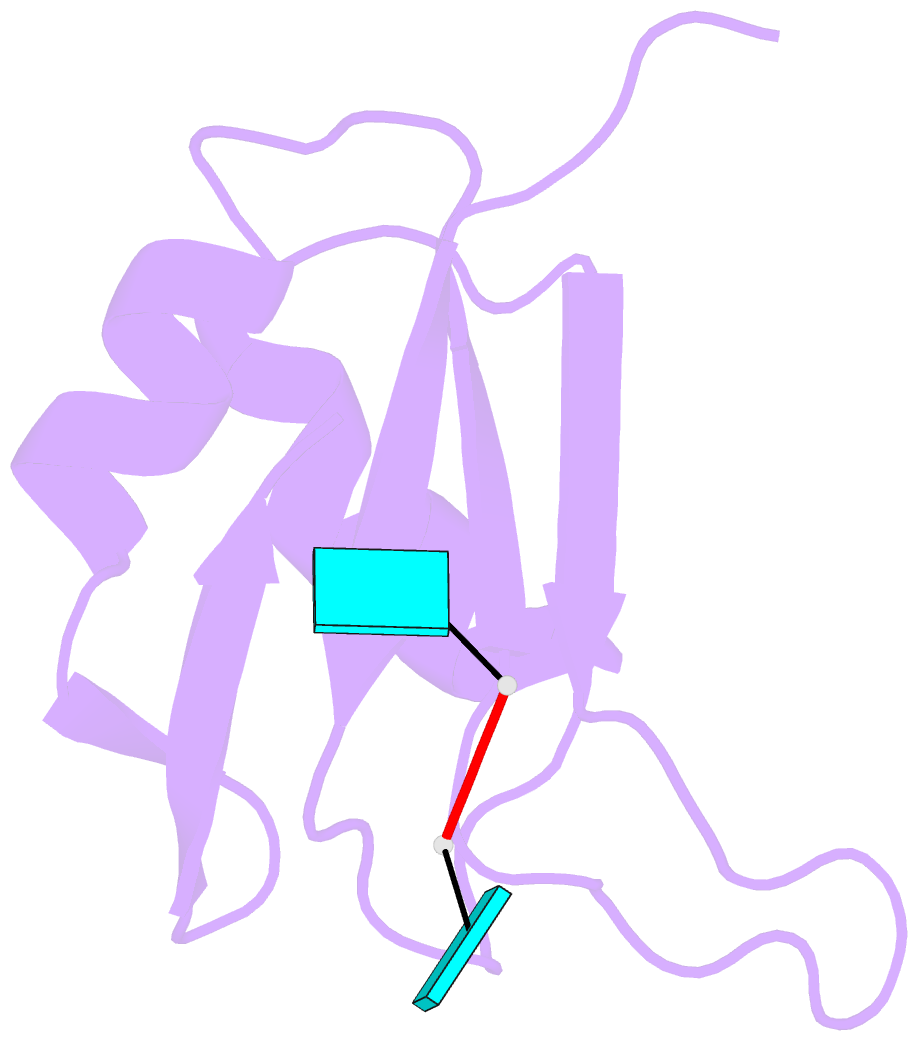
- Crystal structure of tia-1 rrm2 in complex with RNA
- Reference
- Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M (2017): "Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins." Angew. Chem. Int. Ed. Engl., 56, 9322-9325. doi: 10.1002/anie.201702904.
- Abstract
- Multi-domain proteins play critical roles in fine-tuning essential processes in cellular signaling and gene regulation. Typically, multiple globular domains that are connected by flexible linkers undergo dynamic rearrangements upon binding to protein, DNA or RNA ligands. RNA binding proteins (RBPs) represent an important class of multi-domain proteins, which regulate gene expression by recognizing linear or structured RNA sequence motifs. Here, we employ segmental perdeuteration of the three RNA recognition motif (RRM) domains in the RBP TIA-1 using Sortase A mediated protein ligation. We show that domain-selective perdeuteration combined with contrast-matched small-angle neutron scattering (SANS), SAXS and computational modeling provides valuable information to precisely define relative domain arrangements. The approach is generally applicable to study conformational arrangements of individual domains in multi-domain proteins and changes induced by ligand binding.