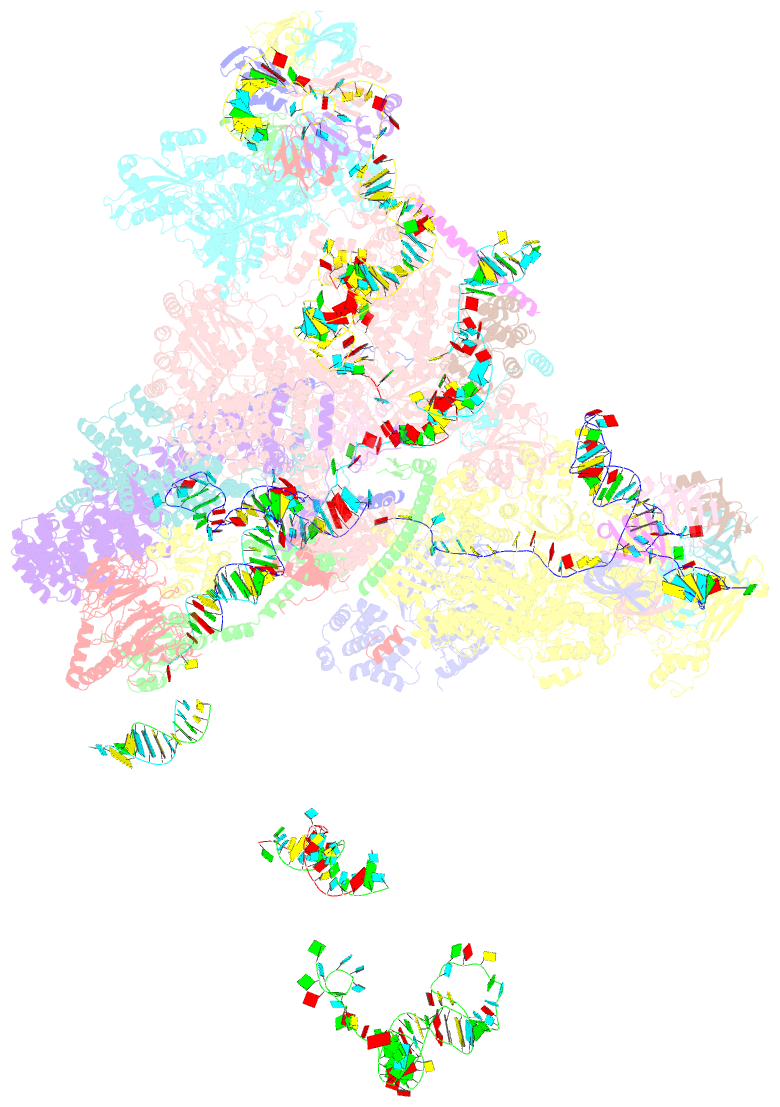
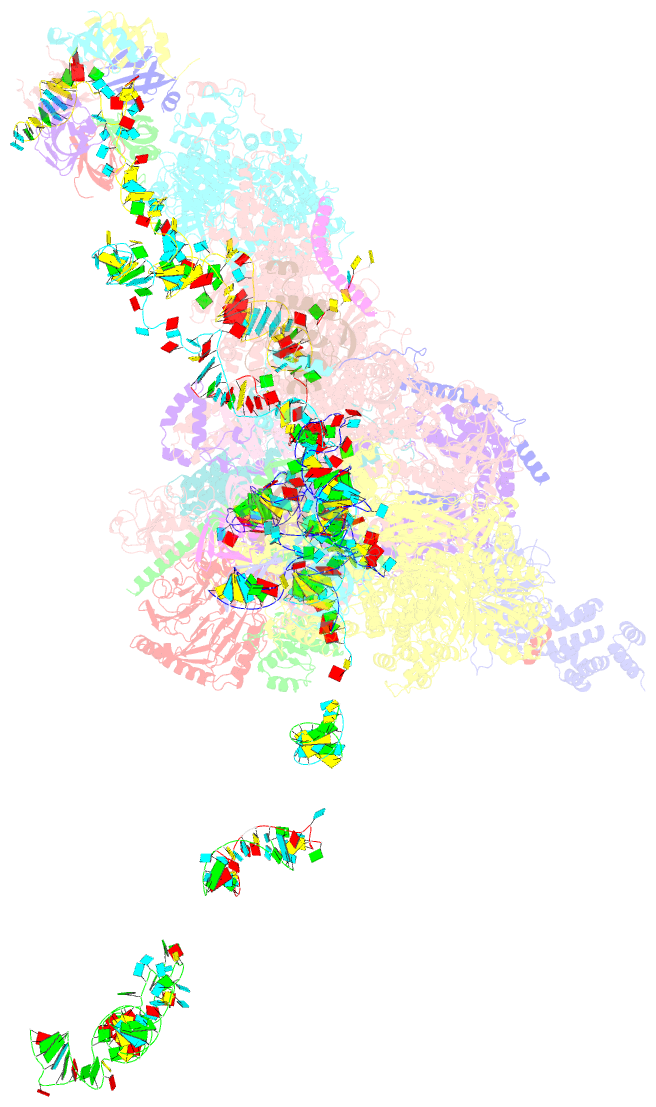
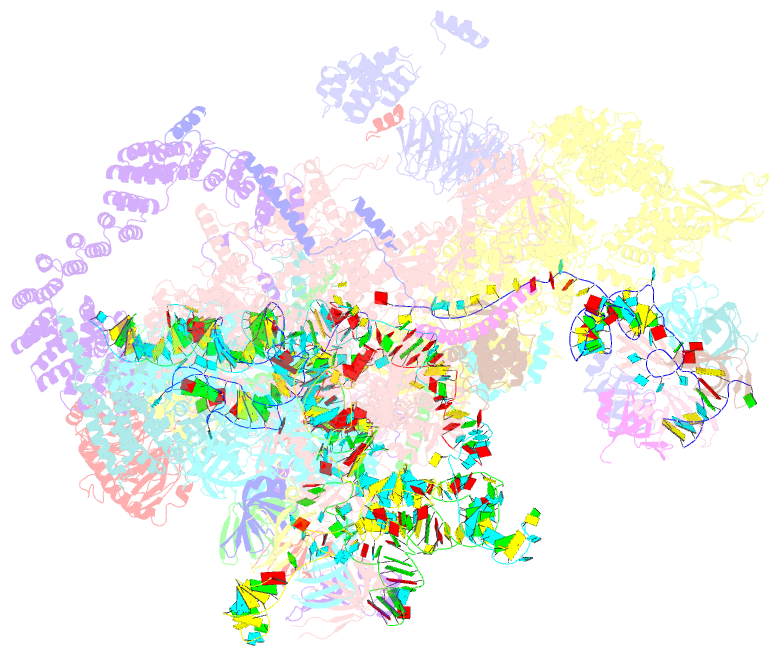
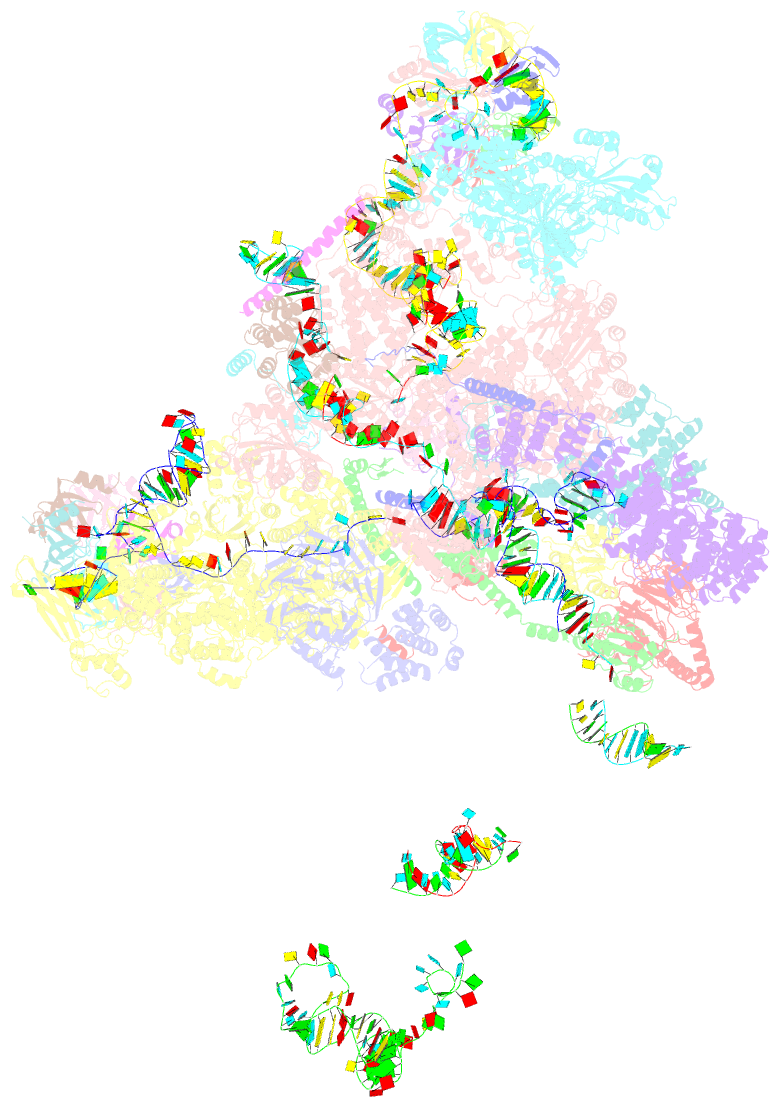
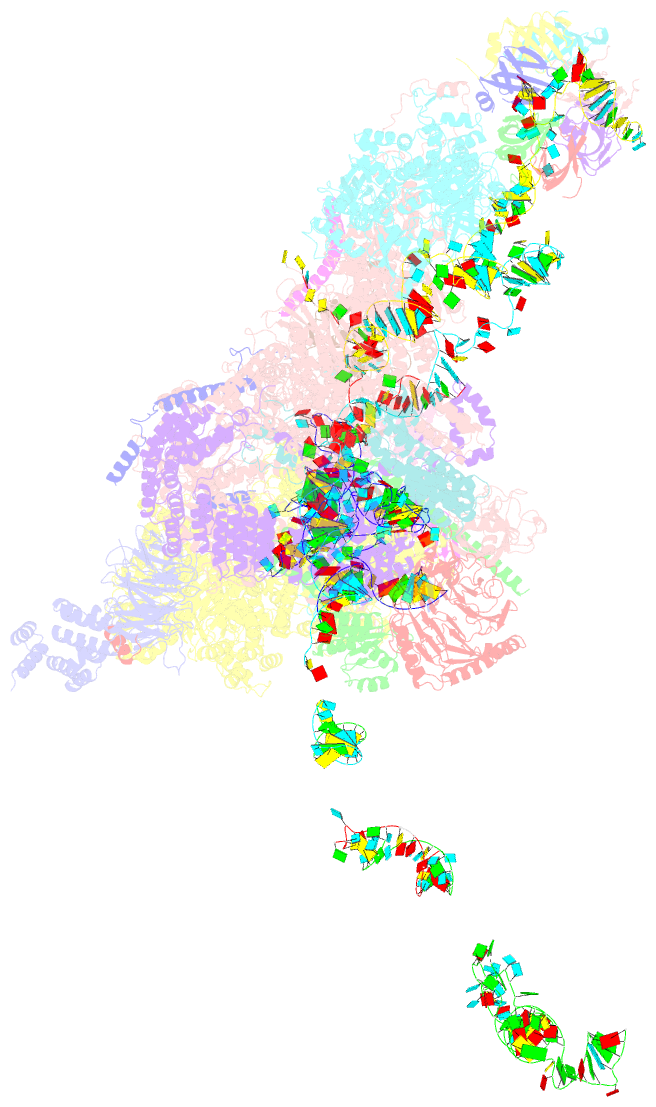
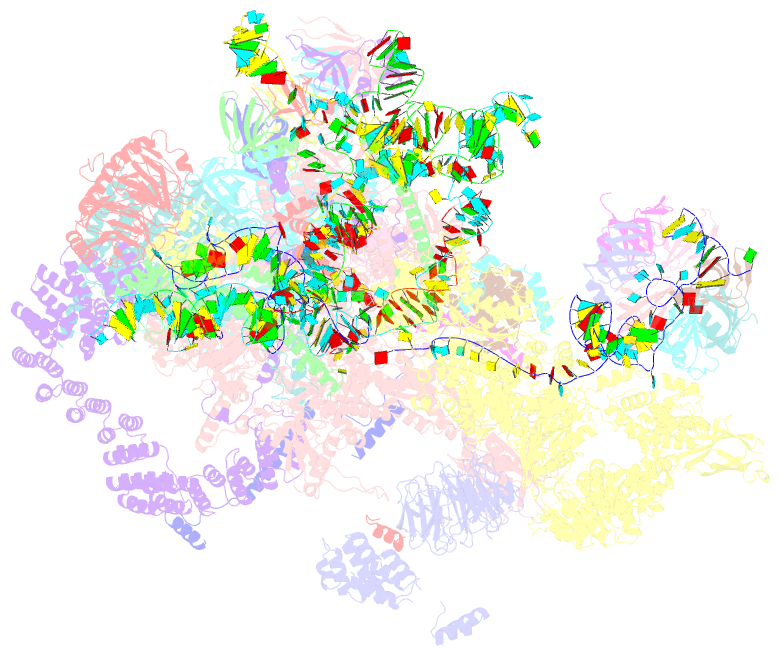
Summary information and primary citation
- PDB-id
- 5o9z; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- cryo-EM (4.5 Å)
- Summary
- cryo-EM structure of a pre-catalytic human spliceosome primed for activation (b complex)
- Reference
- Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Luhrmann R, Stark H (2017): "Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation." Cell, 170, 701-713.e11. doi: 10.1016/j.cell.2017.07.011.
- Abstract
- Little is known about the spliceosome's structure before its extensive remodeling into a catalytically active complex. Here, we report a 3D cryo-EM structure of a pre-catalytic human spliceosomal B complex. The U2 snRNP-containing head domain is connected to the B complex main body via three main bridges. U4/U6.U5 tri-snRNP proteins, which are located in the main body, undergo significant rearrangements during tri-snRNP integration into the B complex. These include formation of a partially closed Prp8 conformation that creates, together with Dim1, a 5' splice site (ss) binding pocket, displacement of Sad1, and rearrangement of Brr2 such that it contacts its U4/U6 substrate and is poised for the subsequent spliceosome activation step. The molecular organization of several B-specific proteins suggests that they are involved in negatively regulating Brr2, positioning the U6/5'ss helix, and stabilizing the B complex structure. Our results indicate significant differences between the early activation phase of human and yeast spliceosomes.